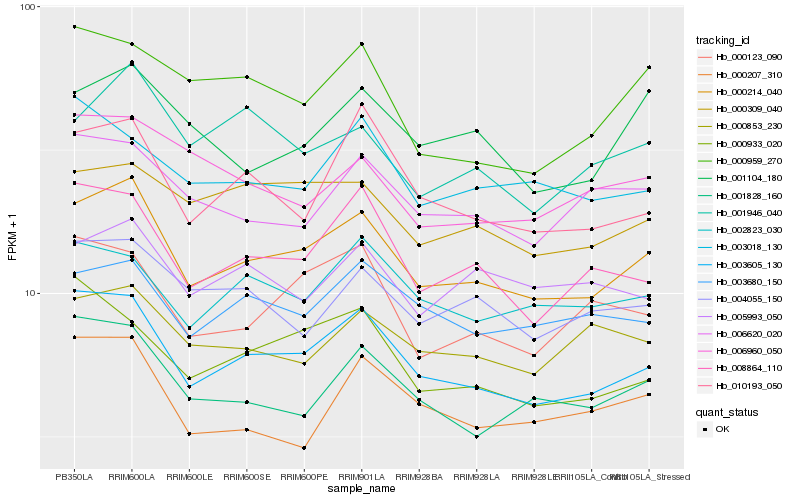
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000214_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003605_130 |
0.0413603999 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |
| 3 |
Hb_008864_110 |
0.0723227541 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 4 |
Hb_003680_150 |
0.077590131 |
- |
- |
srpk, putative [Ricinus communis] |
| 5 |
Hb_001828_160 |
0.0778315371 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 6 |
Hb_010193_050 |
0.0789073871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000309_040 |
0.0793263861 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 8 |
Hb_002823_030 |
0.0803508748 |
- |
- |
- |
| 9 |
Hb_000123_090 |
0.080620263 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 10 |
Hb_004055_150 |
0.0818335256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001104_180 |
0.0824922773 |
- |
- |
PREDICTED: ribosome production factor 2 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_006620_020 |
0.0825217846 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 13 |
Hb_001946_040 |
0.0826571921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000853_230 |
0.0840426373 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_000933_020 |
0.0860233589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005993_050 |
0.0861254358 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 17 |
Hb_003018_130 |
0.0876101952 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 18 |
Hb_006960_050 |
0.0891424416 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 19 |
Hb_000959_270 |
0.0894173958 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 20 |
Hb_000207_310 |
0.090183895 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |