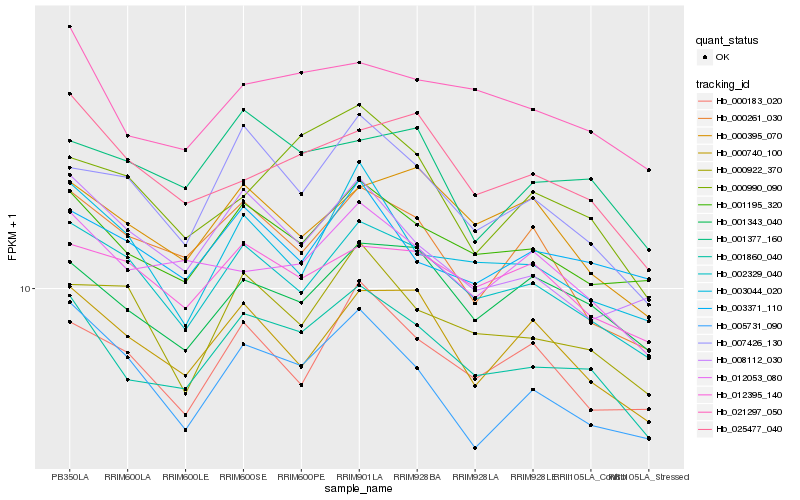
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001860_040 |
0.0 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 2 |
Hb_001195_320 |
0.0622611145 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 3 |
Hb_021297_050 |
0.0664911291 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 4 |
Hb_002329_040 |
0.0678957209 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 5 |
Hb_007426_130 |
0.0743161906 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_001343_040 |
0.0751894872 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 7 |
Hb_012053_080 |
0.0788221328 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 8 |
Hb_000740_100 |
0.0814598083 |
- |
- |
calpain, putative [Ricinus communis] |
| 9 |
Hb_025477_040 |
0.0824907977 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 10 |
Hb_000261_030 |
0.0837957896 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 11 |
Hb_008112_030 |
0.0843323576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000922_370 |
0.084528831 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 13 |
Hb_012395_140 |
0.084639781 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 14 |
Hb_005731_090 |
0.0850067497 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 15 |
Hb_001377_160 |
0.0851799141 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 16 |
Hb_003044_020 |
0.0853427936 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 17 |
Hb_000990_090 |
0.0864948154 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_000183_020 |
0.0865358727 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000395_070 |
0.0870112556 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003371_110 |
0.0870476421 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |