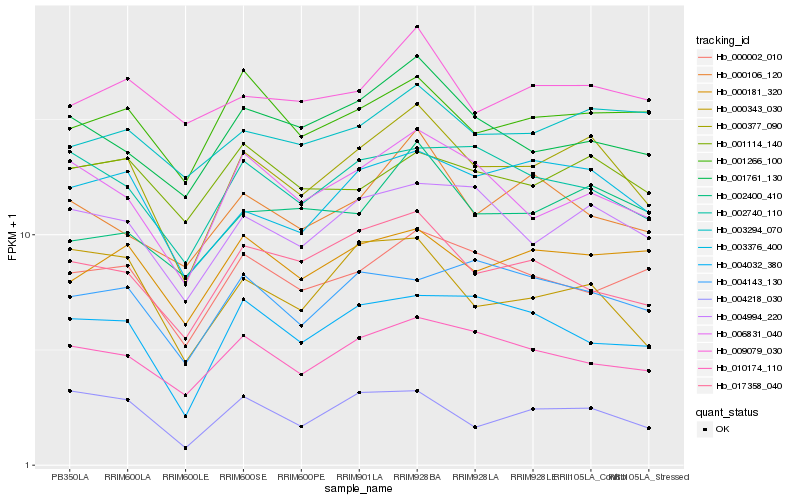
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000377_090 |
0.0 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 2 |
Hb_003376_400 |
0.0858271483 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 3 |
Hb_004994_220 |
0.0866650095 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_006831_040 |
0.0942959655 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_010174_110 |
0.095397526 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 6 |
Hb_002400_410 |
0.0986294014 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 7 |
Hb_017358_040 |
0.1012111865 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 8 |
Hb_001761_130 |
0.1016699777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000181_320 |
0.1020818416 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_004218_030 |
0.1047689706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001266_100 |
0.104917705 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 12 |
Hb_002740_110 |
0.105111178 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 13 |
Hb_000002_010 |
0.1051952777 |
- |
- |
PREDICTED: uncharacterized protein LOC105641567 [Jatropha curcas] |
| 14 |
Hb_000106_120 |
0.106896288 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 15 |
Hb_004032_380 |
0.1076237747 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 16 |
Hb_003294_070 |
0.1081545701 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 17 |
Hb_009079_030 |
0.1087897839 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 18 |
Hb_000343_030 |
0.109311766 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 19 |
Hb_001114_140 |
0.1105826463 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 20 |
Hb_004143_130 |
0.1111997632 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |