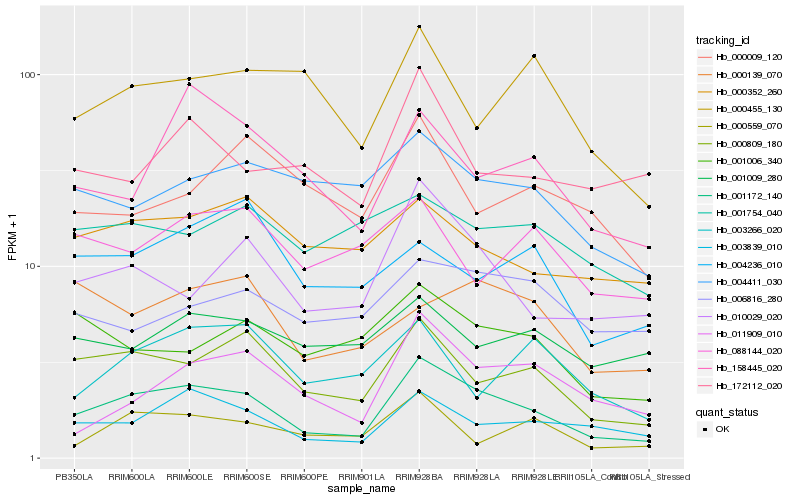
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001172_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 2 |
Hb_000809_180 |
0.1404373982 |
- |
- |
- |
| 3 |
Hb_003839_010 |
0.1515726514 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 4 |
Hb_010029_020 |
0.1596326714 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 5 |
Hb_011909_010 |
0.1814419282 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |
| 6 |
Hb_158445_020 |
0.1818749694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004411_030 |
0.1891485212 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 8 |
Hb_000352_260 |
0.1894141307 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 9 |
Hb_172112_020 |
0.1900048404 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 10 |
Hb_001006_340 |
0.1905730814 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 11 |
Hb_000559_070 |
0.1907175563 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 12 |
Hb_000455_130 |
0.1954304239 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003266_020 |
0.1971933144 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 14 |
Hb_000139_070 |
0.1983079218 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 15 |
Hb_001009_280 |
0.2019987258 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 16 |
Hb_006816_280 |
0.202380053 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001754_040 |
0.203603853 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 18 |
Hb_088144_020 |
0.2038587698 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000009_120 |
0.2046110832 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 20 |
Hb_004236_010 |
0.2049791281 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |