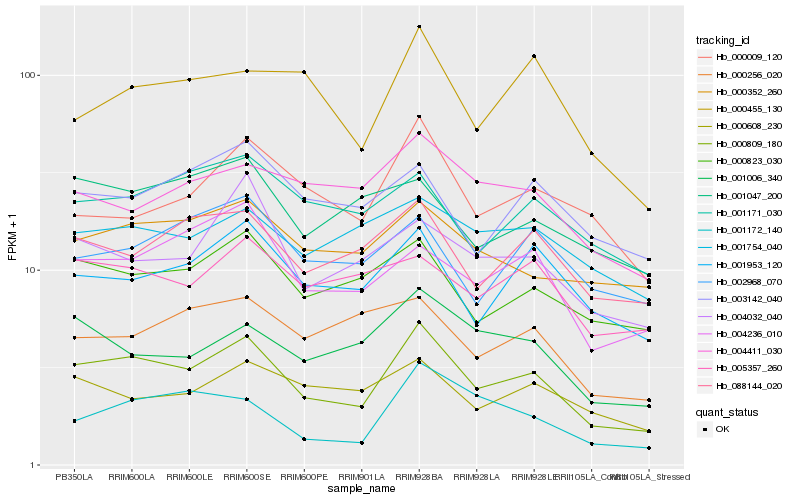
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_000823_030 |
0.1282388905 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 3 |
Hb_001953_120 |
0.1291120319 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004236_010 |
0.1295986572 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 5 |
Hb_088144_020 |
0.131554979 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004032_040 |
0.136673327 |
- |
- |
unknown [Medicago truncatula] |
| 7 |
Hb_001006_340 |
0.1367073179 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 8 |
Hb_000352_260 |
0.1379663062 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 9 |
Hb_001172_140 |
0.1404373982 |
- |
- |
hypothetical protein POPTR_0014s17790g [Populus trichocarpa] |
| 10 |
Hb_000608_230 |
0.1408159981 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000455_130 |
0.1425093041 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000009_120 |
0.1439008819 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 13 |
Hb_001171_030 |
0.1441240937 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 14 |
Hb_005357_260 |
0.1452376441 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 15 |
Hb_003142_040 |
0.1463907294 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 16 |
Hb_002968_070 |
0.1468079761 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 17 |
Hb_004411_030 |
0.1473303342 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 18 |
Hb_001047_200 |
0.1479673534 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 19 |
Hb_000256_020 |
0.1497716881 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 20 |
Hb_001754_040 |
0.1497754902 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |