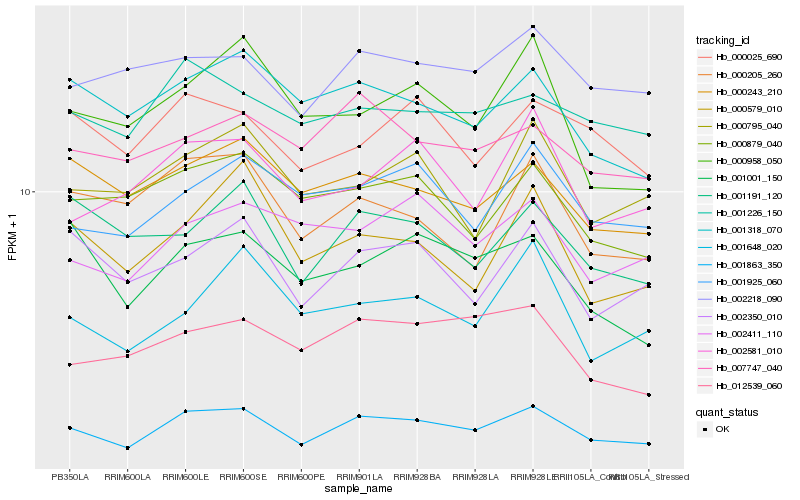
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_350 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 2 |
Hb_002350_010 |
0.0966554495 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 3 |
Hb_001925_060 |
0.0978650229 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 4 |
Hb_001318_070 |
0.0980867265 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001001_150 |
0.1005506122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012539_060 |
0.1014889182 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 7 |
Hb_000958_050 |
0.1017221489 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 8 |
Hb_000243_210 |
0.1021571101 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_002581_010 |
0.1025699089 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000579_010 |
0.1034620731 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 11 |
Hb_002218_090 |
0.1035991196 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000879_040 |
0.1040300128 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_000025_690 |
0.104061737 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 14 |
Hb_001648_020 |
0.1043834846 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 15 |
Hb_002411_110 |
0.1044023373 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 16 |
Hb_001226_150 |
0.1048344719 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_001191_120 |
0.1051467337 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000795_040 |
0.1056541891 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 19 |
Hb_007747_040 |
0.1059951233 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 20 |
Hb_000205_260 |
0.1060803191 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |