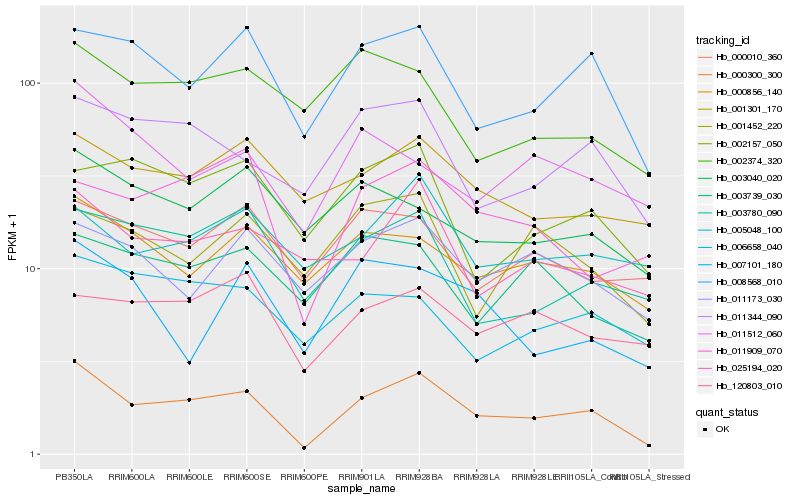
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_300 |
0.0 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_02677 [Jatropha curcas] |
| 2 |
Hb_008568_010 |
0.1572402376 |
- |
- |
Phosphoenolpyruvate carboxykinase [ATP], putative [Ricinus communis] |
| 3 |
Hb_011344_090 |
0.1720015545 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 4 |
Hb_011909_070 |
0.1794525757 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 5 |
Hb_003780_090 |
0.1812359394 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 6 |
Hb_000856_140 |
0.1820686232 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 7 |
Hb_025194_020 |
0.1850515026 |
- |
- |
PREDICTED: uncharacterized protein C1F8.04c-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001301_170 |
0.1852889498 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 9 |
Hb_011173_030 |
0.1880825091 |
- |
- |
pumilio, putative [Ricinus communis] |
| 10 |
Hb_002157_050 |
0.189443625 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 11 |
Hb_006658_040 |
0.1914973909 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 12 |
Hb_011512_060 |
0.1917053535 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_007101_180 |
0.1928630546 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 14 |
Hb_120803_010 |
0.1932578262 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 15 |
Hb_005048_100 |
0.1933967766 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 16 |
Hb_003040_020 |
0.1946712707 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_003739_030 |
0.1949183257 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 18 |
Hb_001452_220 |
0.1958545065 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 19 |
Hb_000010_360 |
0.1961149182 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 20 |
Hb_002374_320 |
0.1967744611 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |