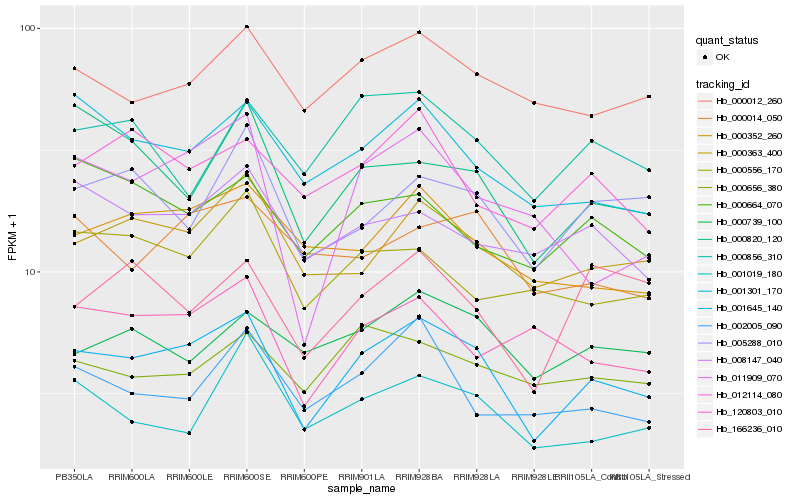
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001645_140 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 2 |
Hb_000363_400 |
0.1106376137 |
- |
- |
PREDICTED: protein HOMOLOG OF MAMMALIAN LYST-INTERACTING PROTEIN 5 [Jatropha curcas] |
| 3 |
Hb_001301_170 |
0.1176402683 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 4 |
Hb_000352_260 |
0.121085661 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 5 |
Hb_000012_260 |
0.1211138807 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 6 |
Hb_005288_010 |
0.1260752663 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_011909_070 |
0.1275542274 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 8 |
Hb_012114_080 |
0.1282347777 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 9 |
Hb_000820_120 |
0.1288389551 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 10 |
Hb_008147_040 |
0.1296799805 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 11 |
Hb_000014_050 |
0.1304387578 |
- |
- |
PREDICTED: V-type proton ATPase subunit B 2 [Elaeis guineensis] |
| 12 |
Hb_000739_100 |
0.1322190924 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 13 |
Hb_120803_010 |
0.1340240326 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 14 |
Hb_001019_180 |
0.1342247893 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 15 |
Hb_000664_070 |
0.1343040122 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 16 |
Hb_002005_090 |
0.1347310713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000656_380 |
0.1351543769 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 18 |
Hb_000856_310 |
0.1352317314 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 19 |
Hb_000556_170 |
0.1362883336 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 20 |
Hb_166236_010 |
0.1371809875 |
- |
- |
- |