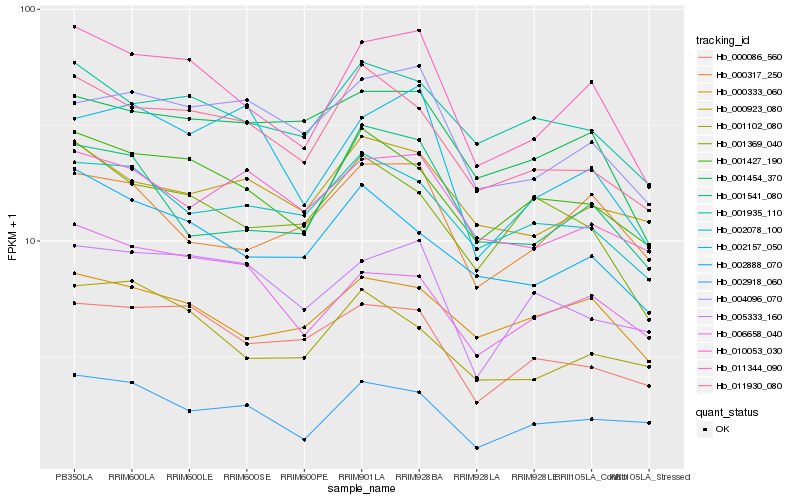
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_090 |
0.0 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 2 |
Hb_001427_190 |
0.0916644215 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 3 |
Hb_011930_080 |
0.0996826439 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 4 |
Hb_002888_070 |
0.1037717655 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 5 |
Hb_000333_060 |
0.1039297373 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 6 |
Hb_001935_110 |
0.1044742357 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 7 |
Hb_000086_560 |
0.1048060514 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004096_070 |
0.10591115 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002918_060 |
0.1073749732 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 10 |
Hb_006658_040 |
0.1087066517 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 11 |
Hb_001541_080 |
0.109241353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000317_250 |
0.1093231708 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001454_370 |
0.1110493391 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 14 |
Hb_002157_050 |
0.1124671142 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 15 |
Hb_002078_100 |
0.1132239392 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 16 |
Hb_005333_160 |
0.1146218172 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 17 |
Hb_001102_080 |
0.1148662174 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 18 |
Hb_000923_080 |
0.1160469813 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 19 |
Hb_001369_040 |
0.1170401709 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010053_030 |
0.1172141398 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |