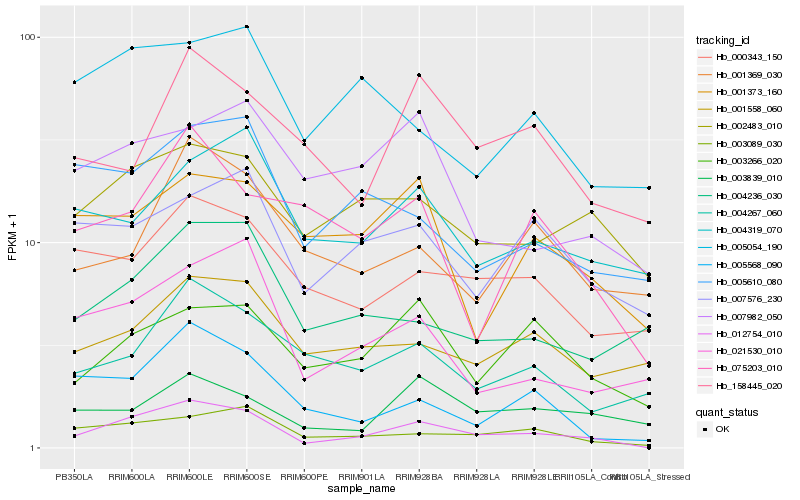
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012754_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003089_030 |
0.1878444547 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 3 |
Hb_005568_090 |
0.2074985346 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_002483_010 |
0.2106215226 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_021530_010 |
0.2116300826 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 6 |
Hb_003266_020 |
0.2148769691 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 7 |
Hb_000343_150 |
0.2173696686 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 8 |
Hb_004267_060 |
0.2197766746 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_004236_030 |
0.2239660701 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_007576_230 |
0.2242740863 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 11 |
Hb_005610_080 |
0.2246443422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001558_060 |
0.2278370956 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 13 |
Hb_075203_010 |
0.2296025623 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004319_070 |
0.2296160804 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 15 |
Hb_001369_030 |
0.2306074064 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 16 |
Hb_158445_020 |
0.2325120719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003839_010 |
0.2325989048 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 18 |
Hb_007982_050 |
0.2333413079 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Jatropha curcas] |
| 19 |
Hb_005054_190 |
0.2342142044 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001373_160 |
0.234371697 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |