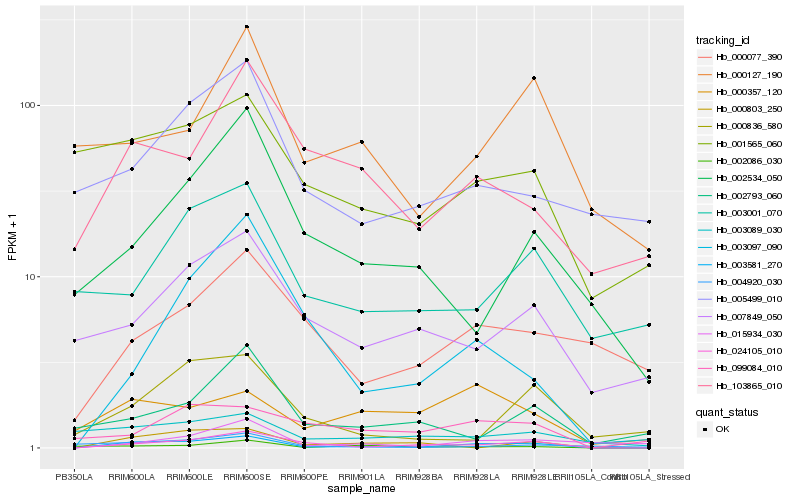
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_270 |
0.0 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_024105_010 |
0.2132000235 |
- |
- |
CCHC-type integrase [Theobroma cacao] |
| 3 |
Hb_000803_250 |
0.2819565031 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015934_030 |
0.2849647125 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_003097_090 |
0.2933975487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002086_030 |
0.2982052358 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 7 |
Hb_000836_580 |
0.3001162899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_103865_010 |
0.3012797094 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 9 |
Hb_007849_050 |
0.3066035998 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 10 |
Hb_003089_030 |
0.3103263078 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001565_060 |
0.3106593787 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002534_050 |
0.3130857616 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 13 |
Hb_000357_120 |
0.3133813639 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 14 |
Hb_099084_010 |
0.314616291 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_000127_190 |
0.314617497 |
- |
- |
PREDICTED: protein GIGANTEA [Jatropha curcas] |
| 16 |
Hb_004920_030 |
0.316697355 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 17 |
Hb_003001_070 |
0.3169276336 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 18 |
Hb_002793_060 |
0.3171067655 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 19 |
Hb_005499_010 |
0.3177250541 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 20 |
Hb_000077_390 |
0.318351103 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0019s13330g [Populus trichocarpa] |