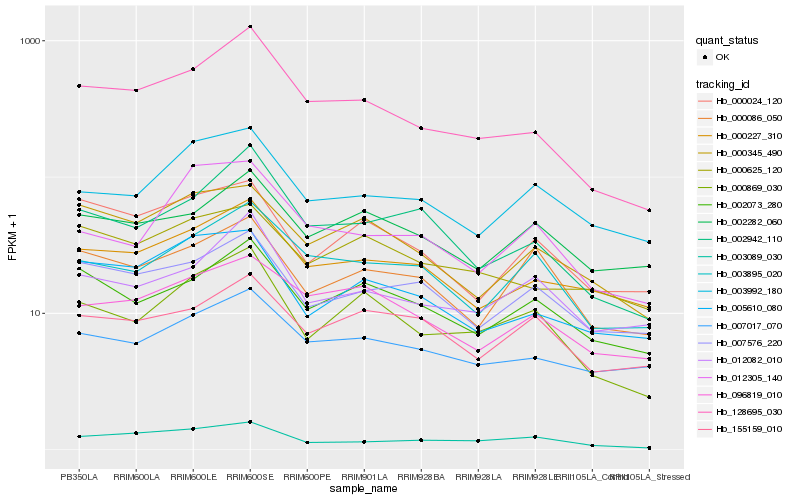
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000869_030 |
0.0 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002073_280 |
0.111534477 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_128695_030 |
0.1139078536 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 4 |
Hb_012082_010 |
0.1212149947 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003895_020 |
0.1249312301 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003992_180 |
0.1256403889 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000086_050 |
0.1265030255 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 8 |
Hb_000345_490 |
0.1303506779 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 9 |
Hb_002942_110 |
0.1315647958 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 10 |
Hb_003089_030 |
0.1318361895 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 11 |
Hb_096819_010 |
0.1331424058 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 12 |
Hb_012305_140 |
0.1332490966 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_155159_010 |
0.1333669588 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 14 |
Hb_002282_060 |
0.1340093425 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000024_120 |
0.1356708929 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 16 |
Hb_005610_080 |
0.1395450545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000227_310 |
0.1403597805 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_007576_220 |
0.1408937391 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 19 |
Hb_000625_120 |
0.141834654 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 20 |
Hb_007017_070 |
0.1424640615 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |