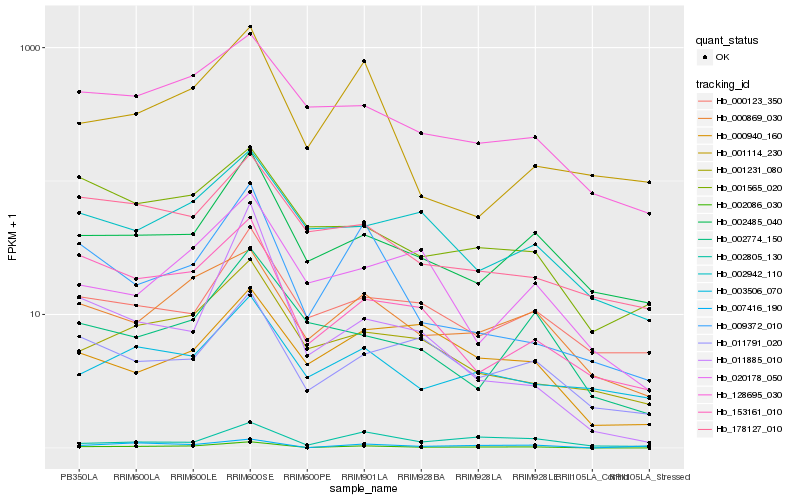
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002086_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_000869_030 |
0.1859264512 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009372_010 |
0.1966292575 |
- |
- |
- |
| 4 |
Hb_128695_030 |
0.1986945364 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 5 |
Hb_001231_080 |
0.2002293202 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_002485_040 |
0.2025353053 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 7 |
Hb_153161_010 |
0.2108293286 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 8 |
Hb_001565_020 |
0.2182051818 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 9 |
Hb_002942_110 |
0.2182818702 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 10 |
Hb_007416_190 |
0.2192561314 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 11 |
Hb_020178_050 |
0.2193223657 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 12 |
Hb_000940_160 |
0.2199090793 |
- |
- |
hypothetical protein VITISV_017596 [Vitis vinifera] |
| 13 |
Hb_002774_150 |
0.2213488134 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 14 |
Hb_011791_020 |
0.2223496159 |
- |
- |
hypothetical protein Csa_6G486930 [Cucumis sativus] |
| 15 |
Hb_002805_130 |
0.2233787709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003506_070 |
0.225747231 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011885_010 |
0.2268902103 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_178127_010 |
0.2281191018 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 19 |
Hb_001114_230 |
0.2294620792 |
- |
- |
- |
| 20 |
Hb_000123_350 |
0.2301942545 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |