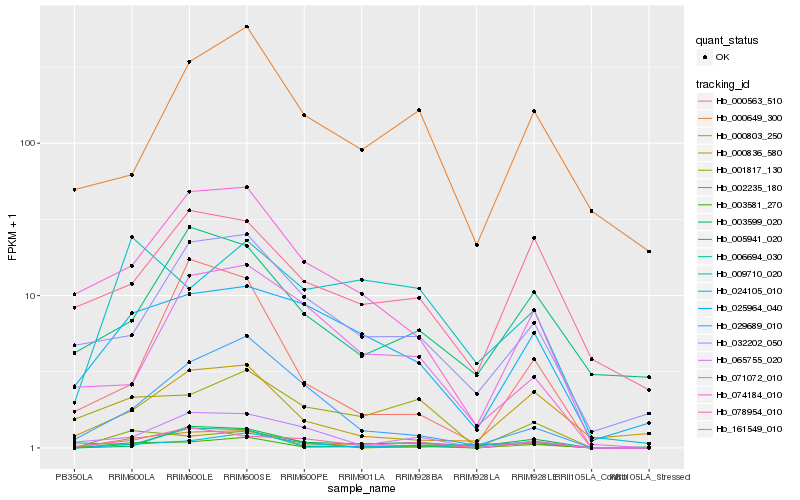
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X1 [Jatropha curcas] |
| 2 |
Hb_078954_010 |
0.2265577197 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 3 |
Hb_025964_040 |
0.2545080843 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 4 |
Hb_003599_020 |
0.2550959314 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_001817_130 |
0.2569535216 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 6 |
Hb_032202_050 |
0.2619894877 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_065755_020 |
0.2696706166 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_000563_510 |
0.272583026 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 9 |
Hb_009710_020 |
0.2740970076 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 10 |
Hb_002235_180 |
0.2761604042 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 11 |
Hb_161549_010 |
0.2764598799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_029689_010 |
0.277620839 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_006694_030 |
0.2782294714 |
- |
- |
- |
| 14 |
Hb_024105_010 |
0.2782583893 |
- |
- |
CCHC-type integrase [Theobroma cacao] |
| 15 |
Hb_005941_020 |
0.280146956 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 16 |
Hb_000649_300 |
0.2803065185 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 17 |
Hb_074184_010 |
0.2810648221 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 18 |
Hb_000836_580 |
0.2812548338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003581_270 |
0.2819565031 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_071072_010 |
0.2821146651 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |