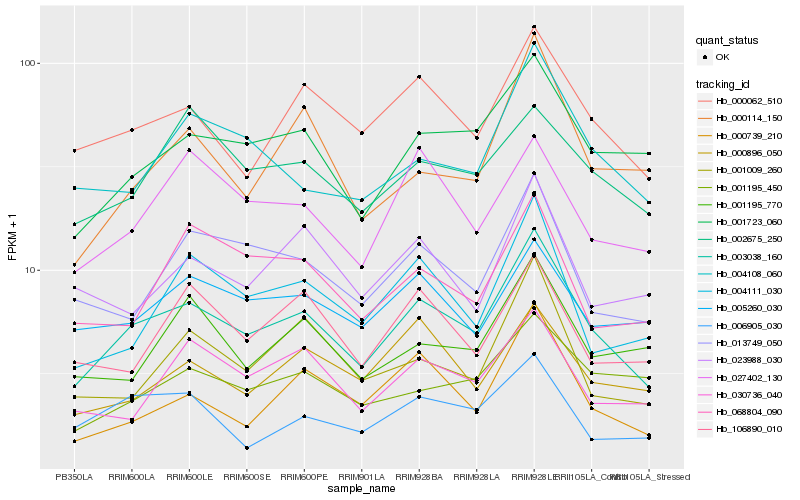
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_160 |
0.0 |
- |
- |
auxilin, putative [Ricinus communis] |
| 2 |
Hb_000062_510 |
0.1055941446 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001723_060 |
0.1126033901 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 4 |
Hb_013749_050 |
0.1199655719 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004111_030 |
0.1211534777 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_004108_060 |
0.1247237867 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 7 |
Hb_030736_040 |
0.1277332432 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000739_210 |
0.1290675086 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 9 |
Hb_001009_260 |
0.1303242016 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005260_030 |
0.1320982811 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 11 |
Hb_027402_130 |
0.1321799071 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000896_050 |
0.1338345046 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 13 |
Hb_001195_770 |
0.1348106569 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 14 |
Hb_106890_010 |
0.1350206524 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_002675_250 |
0.135039179 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 16 |
Hb_000114_150 |
0.1351634796 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 17 |
Hb_023988_030 |
0.1356669114 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 18 |
Hb_068804_090 |
0.1357920995 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 19 |
Hb_001195_450 |
0.1363031044 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 20 |
Hb_006905_030 |
0.1369420616 |
- |
- |
- |