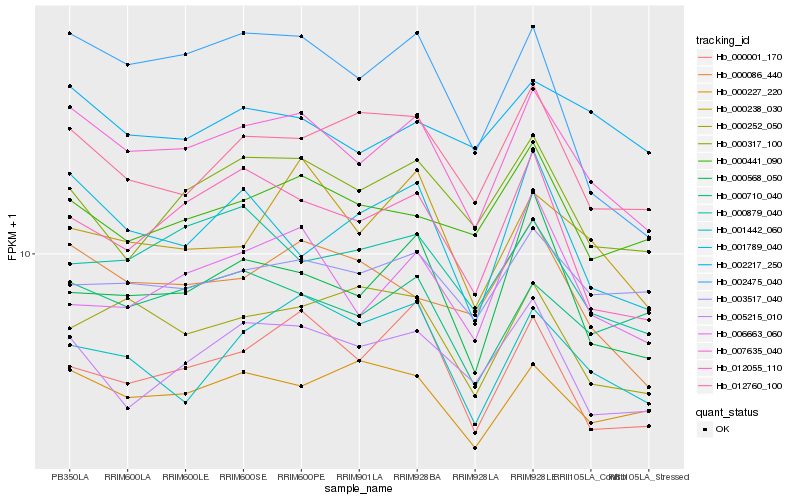
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007635_040 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000086_440 |
0.0744437737 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 3 |
Hb_012055_110 |
0.0752468944 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 4 |
Hb_005215_010 |
0.0767316448 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000710_040 |
0.0767608932 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 6 |
Hb_000317_100 |
0.0776457249 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 7 |
Hb_000568_050 |
0.077693772 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000227_220 |
0.0793021736 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 9 |
Hb_012760_100 |
0.0793312863 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 10 |
Hb_000001_170 |
0.079613191 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_001789_040 |
0.0798563235 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003517_040 |
0.0800894694 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002475_040 |
0.0805074866 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000238_030 |
0.0806347477 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000441_090 |
0.0829047315 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 16 |
Hb_000879_040 |
0.0831693829 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 17 |
Hb_000252_050 |
0.0835961366 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 18 |
Hb_006663_060 |
0.0842216447 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 19 |
Hb_001442_060 |
0.084808542 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002217_250 |
0.085438567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |