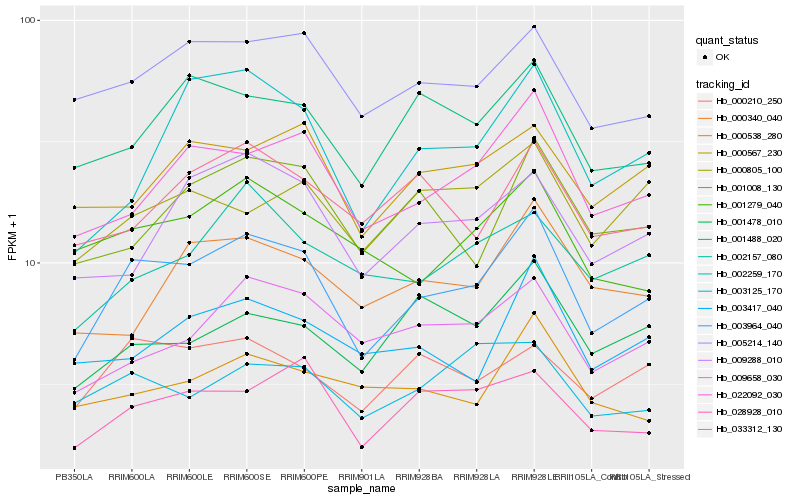
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630528 [Jatropha curcas] |
| 2 |
Hb_022092_030 |
0.1011482518 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_005214_140 |
0.1094737132 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 4 |
Hb_028928_010 |
0.114700298 |
- |
- |
hypothetical protein CISIN_1g0030132mg, partial [Citrus sinensis] |
| 5 |
Hb_000210_250 |
0.1159067124 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X1 [Jatropha curcas] |
| 6 |
Hb_001488_020 |
0.1184593317 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 7 |
Hb_002259_170 |
0.119055404 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_009658_030 |
0.1204767294 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 9 |
Hb_001279_040 |
0.1205329834 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 10 |
Hb_009288_010 |
0.1209907797 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001008_130 |
0.1218317177 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 12 |
Hb_000567_230 |
0.1241988296 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 13 |
Hb_003417_040 |
0.1248036752 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 14 |
Hb_002157_080 |
0.1267543812 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 15 |
Hb_033312_130 |
0.1268332364 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001478_010 |
0.1281974421 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003125_170 |
0.1287184593 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 18 |
Hb_000805_100 |
0.1292984675 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000340_040 |
0.1301022945 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000538_280 |
0.1304084056 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |