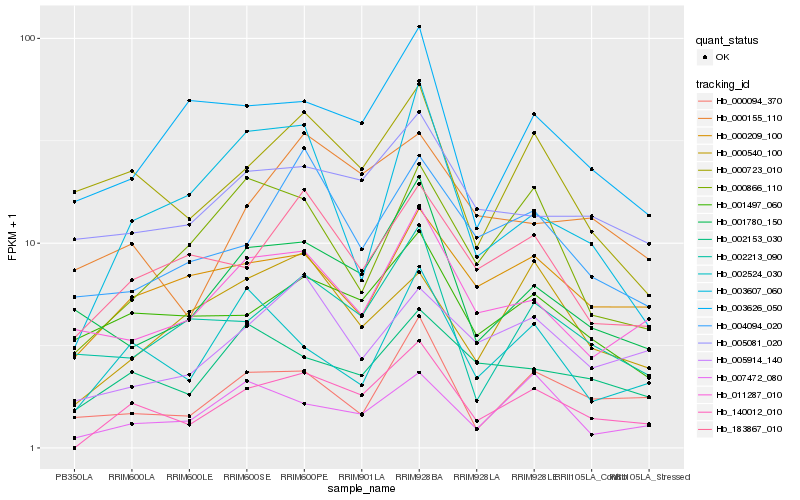
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140012_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Eucalyptus grandis] |
| 2 |
Hb_183867_010 |
0.1894837411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_001497_060 |
0.1963944453 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 4 |
Hb_002153_030 |
0.1978680611 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 5 |
Hb_002524_030 |
0.1978730048 |
- |
- |
- |
| 6 |
Hb_000209_100 |
0.1989181535 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000723_010 |
0.2009134377 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_007472_080 |
0.2009222066 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 9 |
Hb_001780_150 |
0.2012689701 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 10 |
Hb_000094_370 |
0.2016218858 |
- |
- |
- |
| 11 |
Hb_002213_090 |
0.2024640659 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 12 |
Hb_003607_060 |
0.2026115122 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 13 |
Hb_005914_140 |
0.2029370959 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 14 |
Hb_000155_110 |
0.2039125375 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 15 |
Hb_003626_050 |
0.2047707267 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 16 |
Hb_011287_010 |
0.2049532393 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_005081_020 |
0.204968144 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 18 |
Hb_000540_100 |
0.2061065265 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 19 |
Hb_004094_020 |
0.2063496419 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000866_110 |
0.2071652759 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |