| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
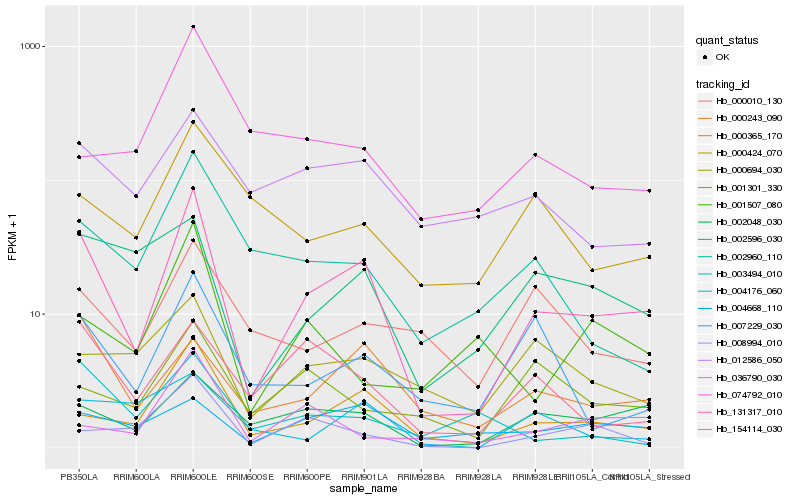
Hb_131317_010 |
0.0 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 2 |
Hb_000365_170 |
0.2011546377 |
- |
- |
PREDICTED: 50S ribosomal protein L17, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_002048_030 |
0.2431159407 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000243_090 |
0.2564197123 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |
| 5 |
Hb_002596_030 |
0.2640588188 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 6 |
Hb_007229_030 |
0.2640608486 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_154114_030 |
0.2676413259 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_002960_110 |
0.2716357706 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012586_050 |
0.2752316749 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 10 |
Hb_008994_010 |
0.2790252922 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_004668_110 |
0.280451055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000694_030 |
0.2847752492 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 13 |
Hb_000424_070 |
0.2879112394 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 14 |
Hb_000010_130 |
0.2896204904 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic [Pyrus x bretschneideri] |
| 15 |
Hb_004176_060 |
0.2902533549 |
- |
- |
- |
| 16 |
Hb_036790_030 |
0.2913188601 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 17 |
Hb_001301_330 |
0.2961478567 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 18 |
Hb_001507_080 |
0.2975731979 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 19 |
Hb_074792_010 |
0.2985482139 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 20 |
Hb_003494_010 |
0.2986190002 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |