| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
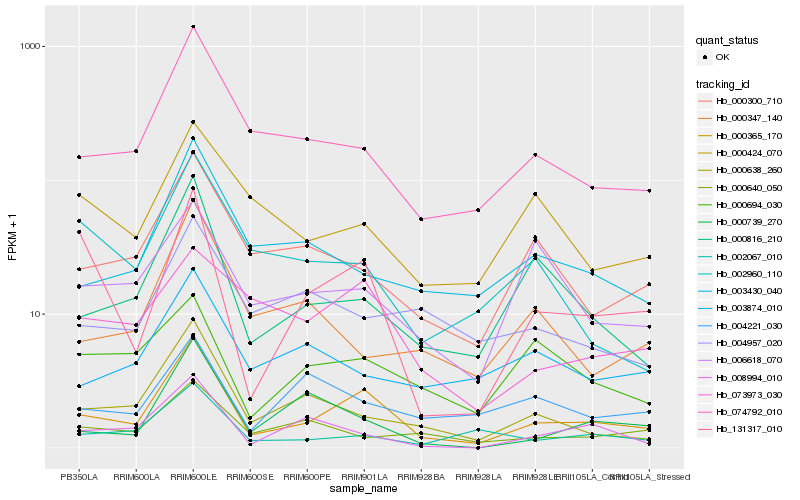
Hb_000365_170 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L17, chloroplastic isoform X3 [Jatropha curcas] |
| 2 |
Hb_074792_010 |
0.1907329554 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 3 |
Hb_000816_210 |
0.1975687442 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 4 |
Hb_131317_010 |
0.2011546377 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_000638_260 |
0.2114436246 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 6 |
Hb_000300_710 |
0.2201727123 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 7 |
Hb_003430_040 |
0.2233903608 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 8 |
Hb_002067_010 |
0.2311128824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008994_010 |
0.2359033209 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000739_270 |
0.2386522457 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 11 |
Hb_073973_030 |
0.2390407534 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000694_030 |
0.2405754626 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 13 |
Hb_002960_110 |
0.2426829326 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000424_070 |
0.2428193993 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 15 |
Hb_006618_070 |
0.2463492303 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003874_010 |
0.2469023028 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000347_140 |
0.2481997764 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_004957_020 |
0.2503295655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000640_050 |
0.2513937761 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 20 |
Hb_004221_030 |
0.2527980163 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |