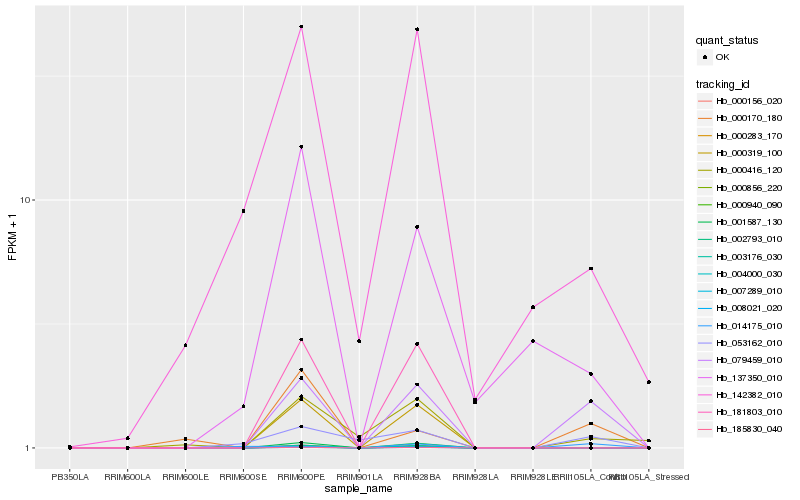
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079459_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_053162_010 |
0.3169973507 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 3 |
Hb_000170_180 |
0.3247925922 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 4 |
Hb_000416_120 |
0.3343807161 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 5 |
Hb_142382_010 |
0.3580094607 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 6 |
Hb_137350_010 |
0.3592206926 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 7 |
Hb_014175_010 |
0.3597511812 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 8 |
Hb_000319_100 |
0.3630690712 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 9 |
Hb_001587_130 |
0.3630722757 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 10 |
Hb_000283_170 |
0.3630756498 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_008021_020 |
0.3630791423 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0014s00780g [Populus trichocarpa] |
| 12 |
Hb_007289_010 |
0.3630809209 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_000156_020 |
0.3630842998 |
- |
- |
PREDICTED: la-related protein 6A [Jatropha curcas] |
| 14 |
Hb_003176_030 |
0.3630842998 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Solanum lycopersicum] |
| 15 |
Hb_000940_090 |
0.3631115418 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 16 |
Hb_000856_220 |
0.3632309523 |
- |
- |
hypothetical protein POPTR_0012s07300g [Populus trichocarpa] |
| 17 |
Hb_002793_010 |
0.3632434353 |
- |
- |
putative metal-nicotianamine transporter YSL7 -like protein [Gossypium arboreum] |
| 18 |
Hb_185830_040 |
0.3632571734 |
- |
- |
hypothetical protein JCGZ_16423 [Jatropha curcas] |
| 19 |
Hb_004000_030 |
0.3632952609 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 20 |
Hb_181803_010 |
0.3632977802 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |