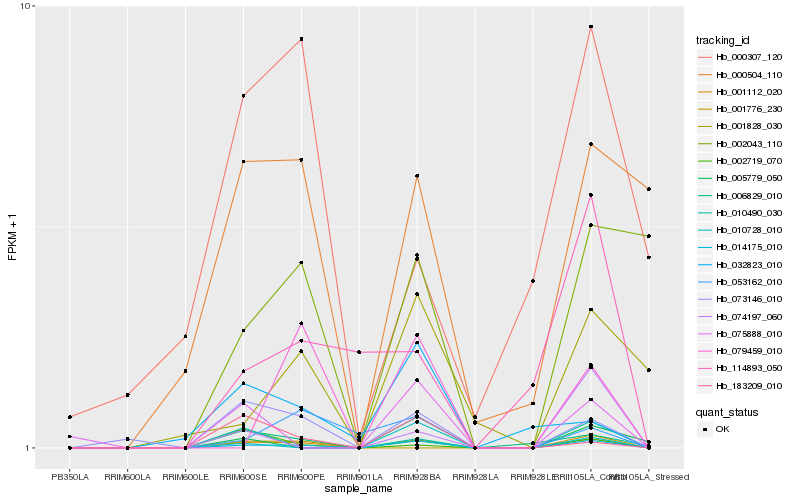
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001112_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 2 |
Hb_014175_010 |
0.09420358 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 3 |
Hb_073146_010 |
0.2703158048 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 4 |
Hb_006829_010 |
0.3110168568 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_010490_030 |
0.3390142676 |
- |
- |
- |
| 6 |
Hb_114893_050 |
0.3555248895 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 7 |
Hb_002719_070 |
0.3561232062 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 8 |
Hb_000504_110 |
0.3639530669 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_074197_060 |
0.3670521329 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 isoform X1 [Populus euphratica] |
| 10 |
Hb_075888_010 |
0.371165556 |
- |
- |
PREDICTED: 25S rRNA (cytosine-C(5))-methyltransferase nop2-like [Solanum lycopersicum] |
| 11 |
Hb_053162_010 |
0.3719471845 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 12 |
Hb_002043_110 |
0.3760432702 |
- |
- |
hypothetical protein POPTR_0001s19500g [Populus trichocarpa] |
| 13 |
Hb_001828_030 |
0.3778451765 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 14 |
Hb_183209_010 |
0.377969024 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 15 |
Hb_079459_010 |
0.3838345292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000307_120 |
0.3843993191 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 17 |
Hb_001776_230 |
0.3924029866 |
- |
- |
PREDICTED: uncharacterized protein LOC104884260 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_032823_010 |
0.3990413136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010728_010 |
0.4009391325 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 20 |
Hb_005779_050 |
0.4012136506 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |