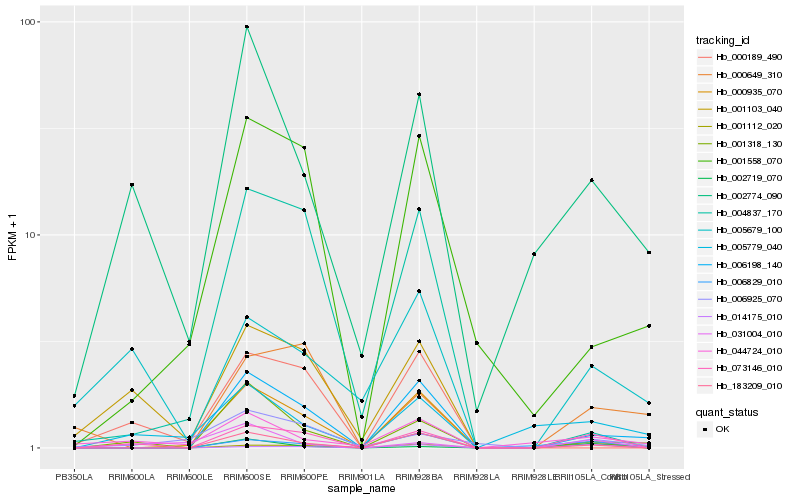
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073146_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 2 |
Hb_001112_020 |
0.2703158048 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 3 |
Hb_183209_010 |
0.273848759 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006198_140 |
0.2978906984 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 5 |
Hb_002774_090 |
0.3000479389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_014175_010 |
0.3014496734 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 7 |
Hb_001318_130 |
0.3043415249 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 8 |
Hb_002719_070 |
0.3170031409 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 9 |
Hb_006829_010 |
0.3223872729 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_001103_040 |
0.3249974579 |
- |
- |
hypothetical protein POPTR_0013s11170g [Populus trichocarpa] |
| 11 |
Hb_005679_100 |
0.3260906587 |
- |
- |
hypothetical protein POPTR_0013s15630g [Populus trichocarpa] |
| 12 |
Hb_000935_070 |
0.3288393773 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000189_490 |
0.3338621905 |
- |
- |
Sulfate/thiosulfate import ATP-binding protein cysA [Theobroma cacao] |
| 14 |
Hb_001558_070 |
0.3360860953 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 15 |
Hb_044724_010 |
0.3370998271 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 16 |
Hb_006925_070 |
0.3390310599 |
- |
- |
hypothetical protein JCGZ_20652 [Jatropha curcas] |
| 17 |
Hb_031004_010 |
0.3396778931 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 18 |
Hb_005779_040 |
0.3402872366 |
- |
- |
- |
| 19 |
Hb_004837_170 |
0.3460202278 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 20 |
Hb_000649_310 |
0.3488206408 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |