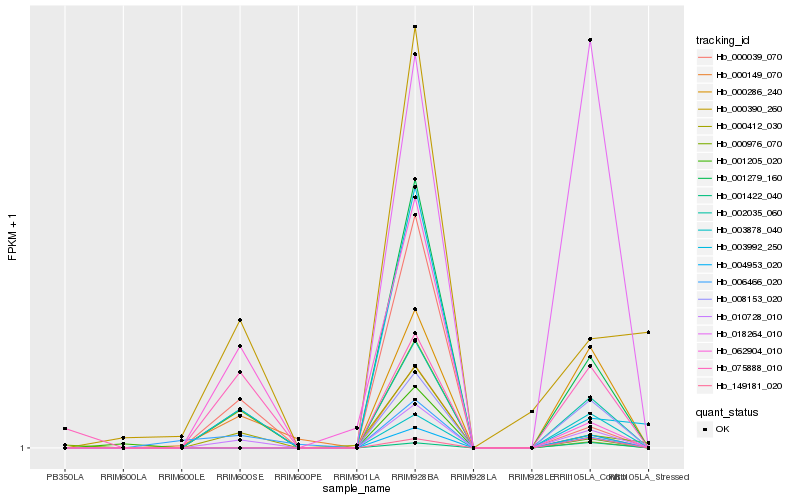
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010728_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 2 |
Hb_002035_060 |
0.2402882841 |
- |
- |
- |
| 3 |
Hb_000039_070 |
0.2443468813 |
transcription factor |
TF Family: MYB |
MYB21, putative [Theobroma cacao] |
| 4 |
Hb_001279_160 |
0.2522720933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008153_020 |
0.2536235936 |
- |
- |
- |
| 6 |
Hb_004953_020 |
0.2567966583 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 7 |
Hb_000412_030 |
0.2621274917 |
- |
- |
glycosyl hydrolase family 35 family protein [Populus trichocarpa] |
| 8 |
Hb_000286_240 |
0.2626763245 |
- |
- |
- |
| 9 |
Hb_075888_010 |
0.2637136036 |
- |
- |
PREDICTED: 25S rRNA (cytosine-C(5))-methyltransferase nop2-like [Solanum lycopersicum] |
| 10 |
Hb_000976_070 |
0.2700801443 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 11 |
Hb_003992_250 |
0.2774196579 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 12 |
Hb_000149_070 |
0.2837488819 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006466_020 |
0.2861499939 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_062904_010 |
0.2945564525 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 15 |
Hb_003878_040 |
0.3059530758 |
- |
- |
- |
| 16 |
Hb_149181_020 |
0.3098154808 |
- |
- |
- |
| 17 |
Hb_018264_010 |
0.3101394907 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 18 |
Hb_001422_040 |
0.3104739349 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 19 |
Hb_001205_020 |
0.3118455914 |
- |
- |
- |
| 20 |
Hb_000390_260 |
0.3163050051 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |