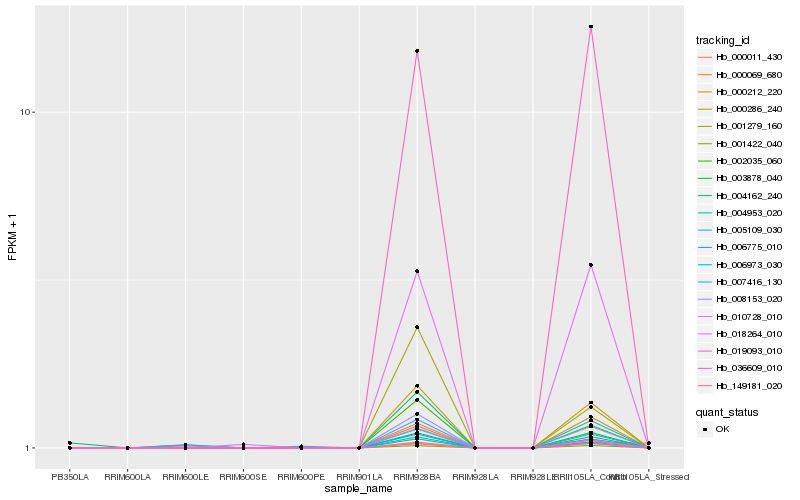
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002035_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_008153_020 |
0.0683260483 |
- |
- |
- |
| 3 |
Hb_004953_020 |
0.0778891296 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 4 |
Hb_000286_240 |
0.0937064681 |
- |
- |
- |
| 5 |
Hb_001279_160 |
0.0979980529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003878_040 |
0.1792026623 |
- |
- |
- |
| 7 |
Hb_149181_020 |
0.1855660683 |
- |
- |
- |
| 8 |
Hb_018264_010 |
0.1860943913 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 9 |
Hb_001422_040 |
0.1866388691 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 10 |
Hb_036609_010 |
0.2101722172 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 11 |
Hb_005109_030 |
0.2221385373 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 12 |
Hb_004162_240 |
0.2351332068 |
transcription factor |
TF Family: C2H2 |
C2H2 and C2HC zinc fingers superfamily protein, putative [Theobroma cacao] |
| 13 |
Hb_010728_010 |
0.2402882841 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 14 |
Hb_000212_220 |
0.2509136213 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 15 |
Hb_006973_030 |
0.2529524314 |
- |
- |
- |
| 16 |
Hb_006775_010 |
0.2544550586 |
transcription factor |
TF Family: M-type |
AGAMOUS [Populus yunnanensis] |
| 17 |
Hb_000069_680 |
0.2668397047 |
- |
- |
- |
| 18 |
Hb_019093_010 |
0.2695462655 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 19 |
Hb_007416_130 |
0.3018130413 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 20 |
Hb_000011_430 |
0.3211271924 |
- |
- |
zinc finger protein, putative [Ricinus communis] |