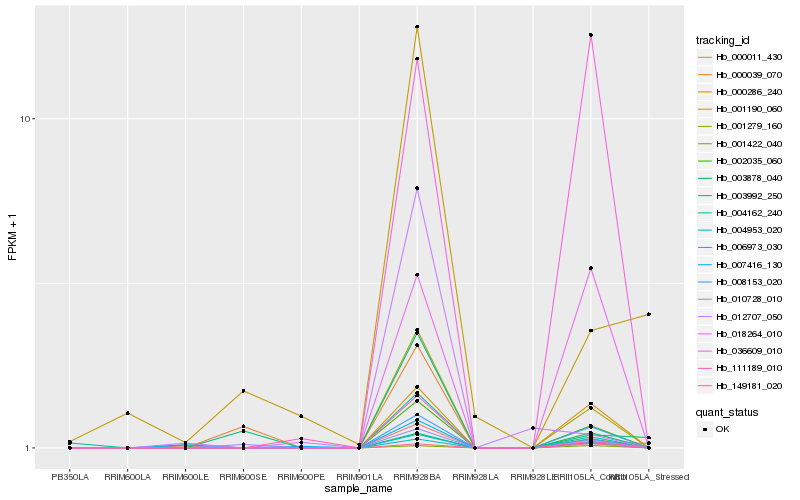
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002035_060 |
0.0979980529 |
- |
- |
- |
| 3 |
Hb_008153_020 |
0.1657760024 |
- |
- |
- |
| 4 |
Hb_004953_020 |
0.1752344212 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 5 |
Hb_004162_240 |
0.1858754914 |
transcription factor |
TF Family: C2H2 |
C2H2 and C2HC zinc fingers superfamily protein, putative [Theobroma cacao] |
| 6 |
Hb_000286_240 |
0.1908652463 |
- |
- |
- |
| 7 |
Hb_010728_010 |
0.2522720933 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 8 |
Hb_007416_130 |
0.2606434845 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 9 |
Hb_003878_040 |
0.2750946211 |
- |
- |
- |
| 10 |
Hb_111189_010 |
0.2807505275 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |
| 11 |
Hb_149181_020 |
0.2813478349 |
- |
- |
- |
| 12 |
Hb_018264_010 |
0.2818669165 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 13 |
Hb_001422_040 |
0.2824018553 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 14 |
Hb_006973_030 |
0.2948911652 |
- |
- |
- |
| 15 |
Hb_012707_050 |
0.2992792473 |
- |
- |
hypothetical protein RCOM_1341590 [Ricinus communis] |
| 16 |
Hb_000011_430 |
0.3022094016 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_036609_010 |
0.3055087023 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 18 |
Hb_003992_250 |
0.3059941334 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 19 |
Hb_001190_060 |
0.3102524922 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 20 |
Hb_000039_070 |
0.3114600283 |
transcription factor |
TF Family: MYB |
MYB21, putative [Theobroma cacao] |