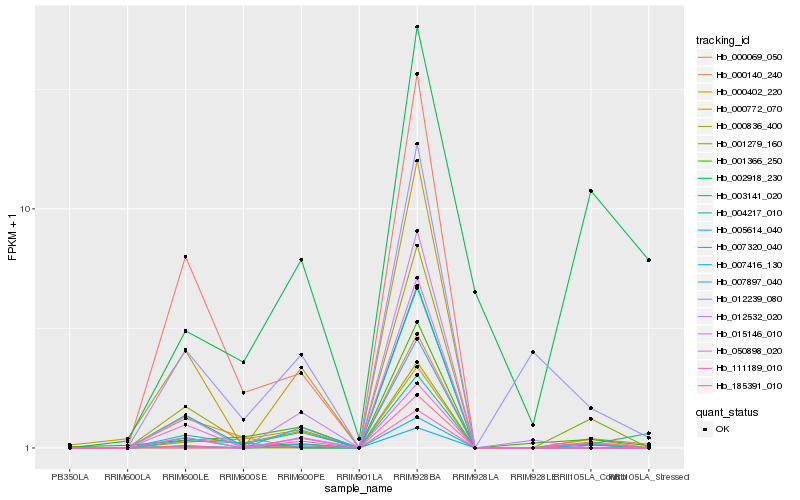
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_130 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 2 |
Hb_007897_040 |
0.2080873451 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_111189_010 |
0.2371379752 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |
| 4 |
Hb_000140_240 |
0.2521484469 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s08640g [Populus trichocarpa] |
| 5 |
Hb_000402_220 |
0.2535000505 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 6 |
Hb_000772_070 |
0.255000244 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 7 |
Hb_000069_050 |
0.2571015336 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 8 |
Hb_000836_400 |
0.2579823501 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 9 |
Hb_015146_010 |
0.2585557655 |
- |
- |
- |
| 10 |
Hb_001279_160 |
0.2606434845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_050898_020 |
0.2609403741 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 12 |
Hb_003141_020 |
0.2613235089 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 13 |
Hb_004217_010 |
0.2626424999 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 14 |
Hb_012532_020 |
0.2650418211 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 15 |
Hb_001366_250 |
0.2654891901 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 16 |
Hb_012239_080 |
0.266136547 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 17 |
Hb_007320_040 |
0.2680306217 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 18 |
Hb_185391_010 |
0.272931044 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 19 |
Hb_005614_040 |
0.2730598789 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_002918_230 |
0.2736124157 |
- |
- |
PREDICTED: probable glutathione S-transferase [Gossypium raimondii] |