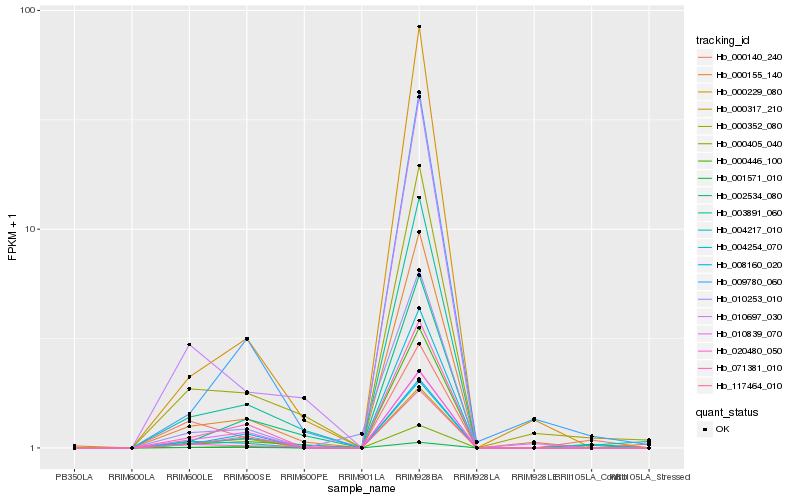
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004217_010 |
0.0 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 2 |
Hb_000140_240 |
0.1047979788 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s08640g [Populus trichocarpa] |
| 3 |
Hb_001571_010 |
0.1247569262 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 4 |
Hb_071381_010 |
0.1422032529 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 5 |
Hb_000352_080 |
0.1477392983 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 6 |
Hb_003891_060 |
0.1505561476 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 7 |
Hb_000405_040 |
0.151028133 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 8 |
Hb_010697_030 |
0.1586840953 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 9 |
Hb_010839_070 |
0.1607716866 |
- |
- |
uncharacterized protein LOC100810725 [Glycine max] |
| 10 |
Hb_020480_050 |
0.1620986077 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 11 |
Hb_000155_140 |
0.1622040796 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000317_210 |
0.1725864317 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39 [Jatropha curcas] |
| 13 |
Hb_009780_060 |
0.1726481891 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_010253_010 |
0.1730567706 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Amborella trichopoda] |
| 15 |
Hb_004254_070 |
0.174226258 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 16 |
Hb_002534_080 |
0.1752265469 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_008160_020 |
0.1768060335 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000446_100 |
0.1771969992 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 19 |
Hb_117464_010 |
0.1773431983 |
- |
- |
PREDICTED: flavone 3'-O-methyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_000229_080 |
0.1775770718 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |