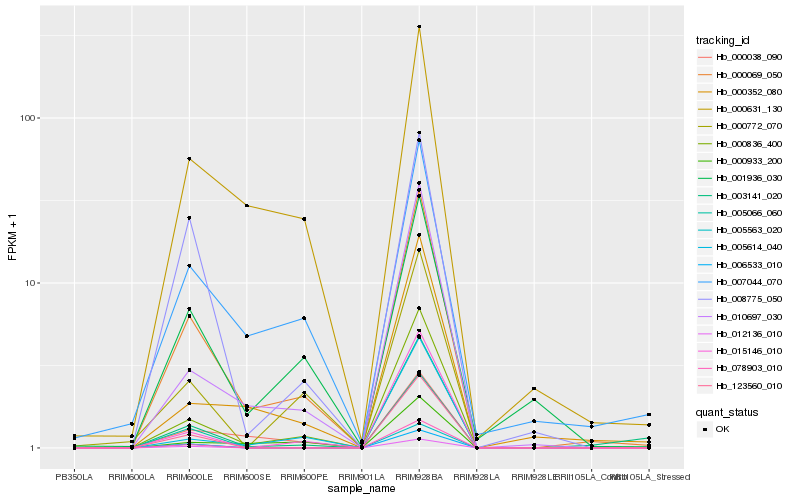
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 2 |
Hb_000836_400 |
0.0917526979 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 3 |
Hb_000631_130 |
0.1278107965 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 4 |
Hb_000038_090 |
0.1299667022 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 5 |
Hb_007044_070 |
0.1316458973 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 6 |
Hb_010697_030 |
0.1328679838 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 7 |
Hb_008775_050 |
0.1424322521 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 8 |
Hb_001936_030 |
0.1441458083 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 9 |
Hb_015146_010 |
0.1449712336 |
- |
- |
- |
| 10 |
Hb_000772_070 |
0.1498362666 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 11 |
Hb_005563_020 |
0.150009277 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 12 |
Hb_012136_010 |
0.1509285004 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 13 |
Hb_005066_060 |
0.1509987723 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 14 |
Hb_000933_200 |
0.151063114 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_123560_010 |
0.1533062736 |
- |
- |
hypothetical protein JCGZ_12463 [Jatropha curcas] |
| 16 |
Hb_006533_010 |
0.1545065654 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 17 |
Hb_003141_020 |
0.1568641602 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 18 |
Hb_000352_080 |
0.1572949458 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 19 |
Hb_078903_010 |
0.1581861729 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 20 |
Hb_005614_040 |
0.1585855542 |
- |
- |
unnamed protein product [Coffea canephora] |