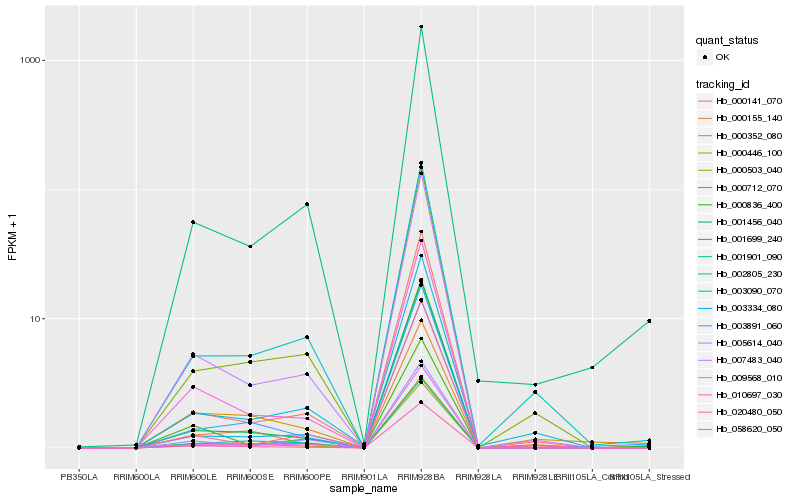
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010697_030 |
0.0 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 2 |
Hb_007483_040 |
0.0385458857 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 3 |
Hb_020480_050 |
0.0518087525 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 4 |
Hb_000503_040 |
0.0704657752 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 5 |
Hb_000836_400 |
0.0741181816 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 6 |
Hb_000141_070 |
0.0770437743 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_009568_010 |
0.0774958114 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 8 |
Hb_005614_040 |
0.0788062482 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000712_070 |
0.0819146072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001456_040 |
0.0823902868 |
- |
- |
NAD dependent epimerase/dehydratase, putative [Ricinus communis] |
| 11 |
Hb_000446_100 |
0.0845897158 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 12 |
Hb_003090_070 |
0.0870121738 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 13 |
Hb_003334_080 |
0.0897328187 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 14 |
Hb_001699_240 |
0.092409483 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 15 |
Hb_058620_050 |
0.0928557371 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_003891_060 |
0.0949642317 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 17 |
Hb_002805_230 |
0.0952628577 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000155_140 |
0.0966308458 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001901_090 |
0.0969361258 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000352_080 |
0.0972873712 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |