| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
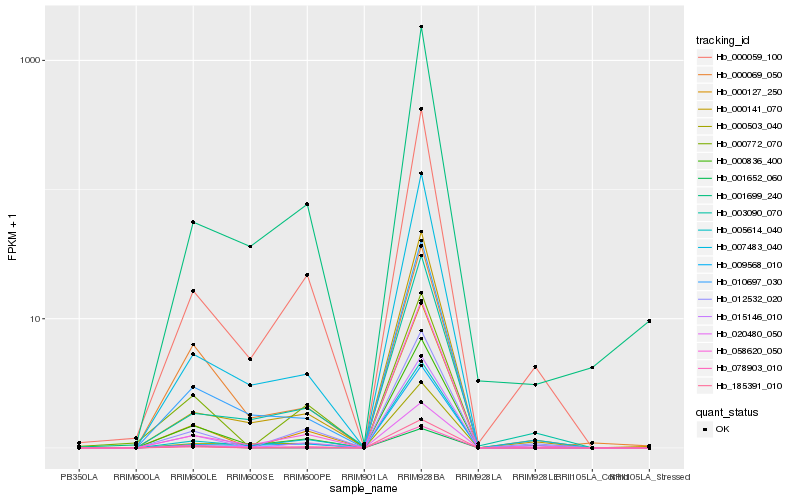
Hb_000836_400 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 1 [Jatropha curcas] |
| 2 |
Hb_015146_010 |
0.0698570764 |
- |
- |
- |
| 3 |
Hb_010697_030 |
0.0741181816 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 4 |
Hb_005614_040 |
0.0826148589 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000069_050 |
0.0917526979 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 6 |
Hb_007483_040 |
0.0919404544 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 7 |
Hb_000059_100 |
0.1057422547 |
- |
- |
Basic blue protein, putative [Ricinus communis] |
| 8 |
Hb_000503_040 |
0.1077534166 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 9 |
Hb_020480_050 |
0.1077607106 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 10 |
Hb_185391_010 |
0.1099646587 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 11 |
Hb_000127_250 |
0.1162518726 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 12 |
Hb_001699_240 |
0.1186087108 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 13 |
Hb_012532_020 |
0.1190281941 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 14 |
Hb_009568_010 |
0.1210971581 |
- |
- |
hypothetical protein POPTR_0001s46700g [Populus trichocarpa] |
| 15 |
Hb_000772_070 |
0.1211597057 |
- |
- |
PREDICTED: uncharacterized protein LOC105649697 [Jatropha curcas] |
| 16 |
Hb_058620_050 |
0.1229034726 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 17 |
Hb_000141_070 |
0.1258386327 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_003090_070 |
0.1275974055 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 19 |
Hb_001652_060 |
0.1304305829 |
- |
- |
PREDICTED: uncharacterized protein LOC103404034 [Malus domestica] |
| 20 |
Hb_078903_010 |
0.1326030134 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |