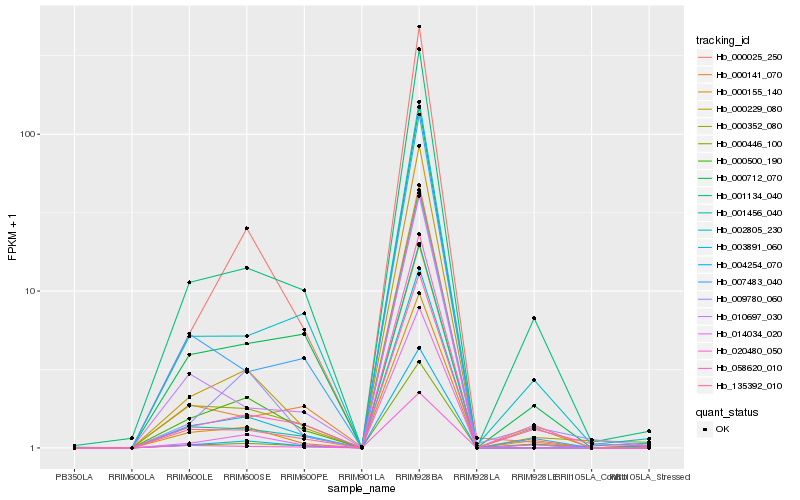
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_140 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_003891_060 |
0.072884031 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 3 |
Hb_000229_080 |
0.0765871961 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 4 |
Hb_000352_080 |
0.0786132304 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 5 |
Hb_001134_040 |
0.0797630745 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 6 |
Hb_058620_010 |
0.0812857655 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_009780_060 |
0.0843286194 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_000500_190 |
0.0845238884 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 9 |
Hb_004254_070 |
0.0878358851 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 10 |
Hb_001456_040 |
0.0896751381 |
- |
- |
NAD dependent epimerase/dehydratase, putative [Ricinus communis] |
| 11 |
Hb_000446_100 |
0.0903183933 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 12 |
Hb_020480_050 |
0.0936332989 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 13 |
Hb_000712_070 |
0.0945118322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_014034_020 |
0.0947887522 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 15 |
Hb_000025_250 |
0.0965964867 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 16 |
Hb_010697_030 |
0.0966308458 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 17 |
Hb_000141_070 |
0.0994865908 |
- |
- |
l-lactate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_007483_040 |
0.1001500332 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 19 |
Hb_135392_010 |
0.100554315 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_002805_230 |
0.1020097471 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |