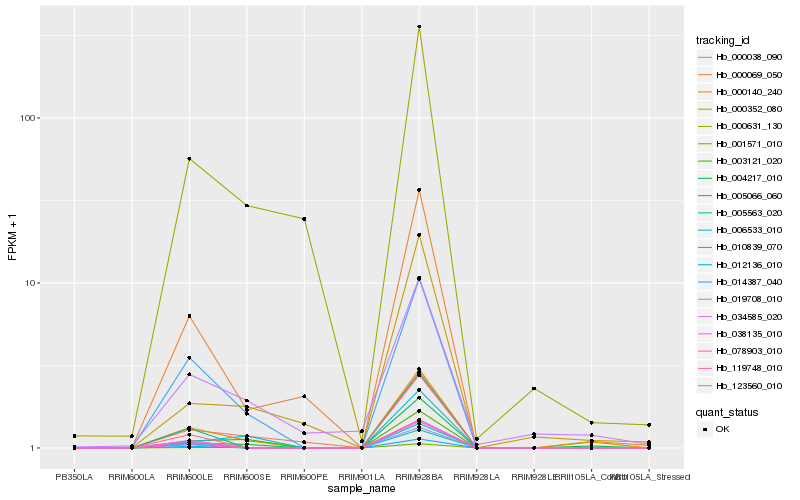
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_240 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s08640g [Populus trichocarpa] |
| 2 |
Hb_004217_010 |
0.1047979788 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 3 |
Hb_014387_040 |
0.1509156833 |
- |
- |
PREDICTED: uncharacterized protein LOC105636953 [Jatropha curcas] |
| 4 |
Hb_001571_010 |
0.1656589277 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 5 |
Hb_000069_050 |
0.1726888715 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 6 |
Hb_000038_090 |
0.1937478145 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 7 |
Hb_003121_020 |
0.1940021516 |
- |
- |
secretory peroxidase PX3 [Manihot esculenta] |
| 8 |
Hb_034585_020 |
0.1945120317 |
- |
- |
- |
| 9 |
Hb_012136_010 |
0.1992742382 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 10 |
Hb_005066_060 |
0.1992908992 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 11 |
Hb_006533_010 |
0.2006839286 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 12 |
Hb_005563_020 |
0.200823503 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 13 |
Hb_010839_070 |
0.2030442434 |
- |
- |
uncharacterized protein LOC100810725 [Glycine max] |
| 14 |
Hb_119748_010 |
0.203754532 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 15 |
Hb_123560_010 |
0.2047410185 |
- |
- |
hypothetical protein JCGZ_12463 [Jatropha curcas] |
| 16 |
Hb_019708_010 |
0.2082155902 |
- |
- |
- |
| 17 |
Hb_000631_130 |
0.2087603593 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 18 |
Hb_000352_080 |
0.2090304845 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 19 |
Hb_038135_010 |
0.2094002874 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 20 |
Hb_078903_010 |
0.2095302456 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |