| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
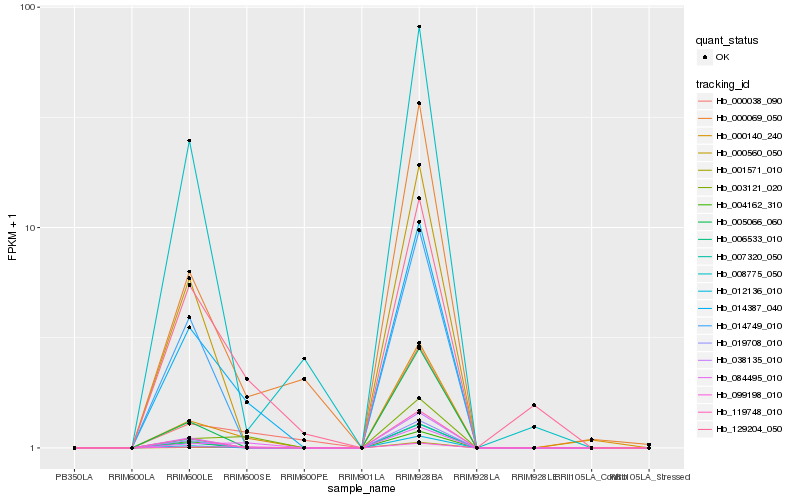
Hb_014387_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636953 [Jatropha curcas] |
| 2 |
Hb_129204_050 |
0.1502529666 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 330-like [Prunus mume] |
| 3 |
Hb_000140_240 |
0.1509156833 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s08640g [Populus trichocarpa] |
| 4 |
Hb_000560_050 |
0.1566253317 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 5 |
Hb_038135_010 |
0.1568686801 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 6 |
Hb_019708_010 |
0.157155999 |
- |
- |
- |
| 7 |
Hb_003121_020 |
0.1592705056 |
- |
- |
secretory peroxidase PX3 [Manihot esculenta] |
| 8 |
Hb_119748_010 |
0.1596761072 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_014749_010 |
0.1622761914 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 10 |
Hb_001571_010 |
0.1627242302 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 11 |
Hb_000038_090 |
0.1630658897 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 12 |
Hb_099198_010 |
0.1642375576 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 13 |
Hb_006533_010 |
0.164318752 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 14 |
Hb_007320_050 |
0.1643953882 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 15 |
Hb_000069_050 |
0.1654848984 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 16 |
Hb_004162_310 |
0.1664052073 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 17 |
Hb_084495_010 |
0.166698293 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Eucalyptus grandis] |
| 18 |
Hb_008775_050 |
0.1684433343 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 19 |
Hb_005066_060 |
0.1705976841 |
- |
- |
FAD-binding Berberine family protein, putative [Theobroma cacao] |
| 20 |
Hb_012136_010 |
0.1708013905 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |