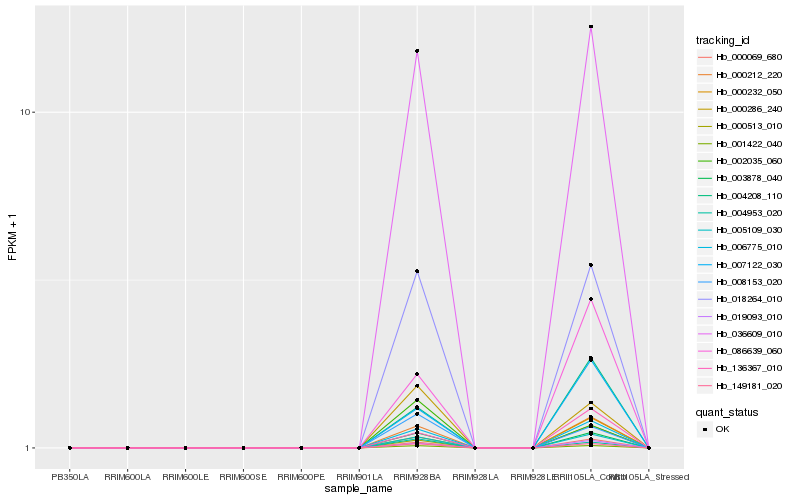
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018264_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 2 |
Hb_149181_020 |
0.0005349012 |
- |
- |
- |
| 3 |
Hb_001422_040 |
0.000551294 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 4 |
Hb_003878_040 |
0.0069747565 |
- |
- |
- |
| 5 |
Hb_036609_010 |
0.0244151419 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 6 |
Hb_005109_030 |
0.0365764414 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 7 |
Hb_000212_220 |
0.0658952219 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 8 |
Hb_006775_010 |
0.0695109445 |
transcription factor |
TF Family: M-type |
AGAMOUS [Populus yunnanensis] |
| 9 |
Hb_000069_680 |
0.0821682331 |
- |
- |
- |
| 10 |
Hb_019093_010 |
0.0849370579 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 11 |
Hb_000286_240 |
0.0929910633 |
- |
- |
- |
| 12 |
Hb_004953_020 |
0.1087974141 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 13 |
Hb_008153_020 |
0.1183367542 |
- |
- |
- |
| 14 |
Hb_004208_110 |
0.1859410808 |
- |
- |
- |
| 15 |
Hb_007122_030 |
0.1860569473 |
- |
- |
- |
| 16 |
Hb_002035_060 |
0.1860943913 |
- |
- |
- |
| 17 |
Hb_086639_060 |
0.1882055912 |
- |
- |
hypothetical protein JCGZ_25622 [Jatropha curcas] |
| 18 |
Hb_136367_010 |
0.194652308 |
- |
- |
hypothetical protein POPTR_0012s02040g [Populus trichocarpa] |
| 19 |
Hb_000232_050 |
0.2030515788 |
- |
- |
- |
| 20 |
Hb_000513_010 |
0.2088919614 |
- |
- |
- |