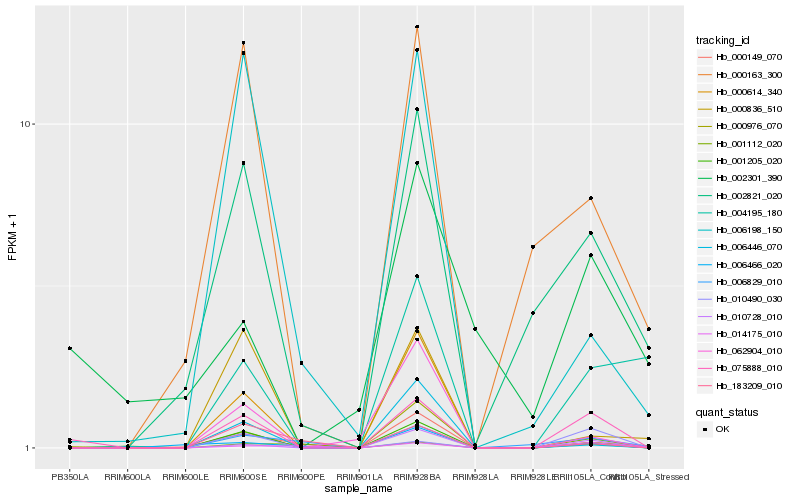
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075888_010 |
0.0 |
- |
- |
PREDICTED: 25S rRNA (cytosine-C(5))-methyltransferase nop2-like [Solanum lycopersicum] |
| 2 |
Hb_010728_010 |
0.2637136036 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 3 |
Hb_010490_030 |
0.3122800718 |
- |
- |
- |
| 4 |
Hb_000976_070 |
0.3131161255 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 5 |
Hb_001205_020 |
0.3194803932 |
- |
- |
- |
| 6 |
Hb_002821_020 |
0.3384654612 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 7 |
Hb_014175_010 |
0.3424360952 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 8 |
Hb_000149_070 |
0.3459893161 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000163_300 |
0.3539331714 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 10 |
Hb_006829_010 |
0.3682828692 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_001112_020 |
0.371165556 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 12 |
Hb_006466_020 |
0.3714356066 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_062904_010 |
0.3737784065 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 14 |
Hb_004195_180 |
0.3767499212 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_006198_150 |
0.3819605344 |
- |
- |
- |
| 16 |
Hb_002301_390 |
0.3836369364 |
- |
- |
- |
| 17 |
Hb_000836_510 |
0.385050778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_183209_010 |
0.3888649389 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000614_340 |
0.3931843922 |
- |
- |
Salutaridinol 7-O-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_006446_070 |
0.3952823073 |
- |
- |
Zinc-binding dehydrogenase family protein [Theobroma cacao] |