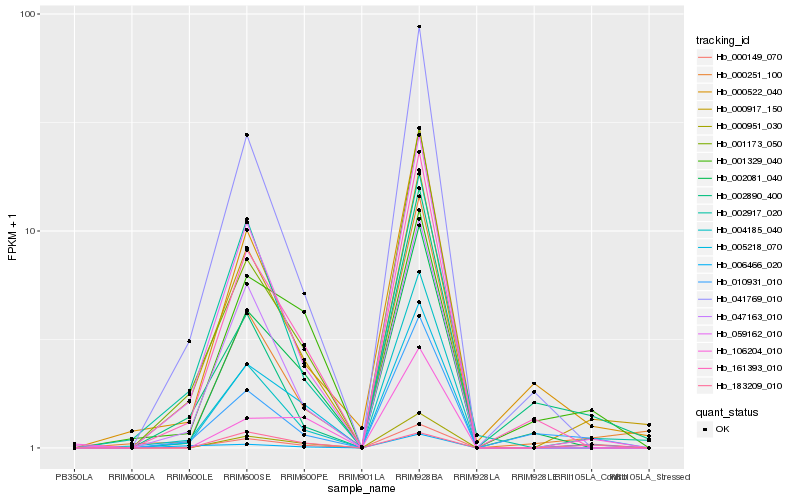
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000149_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000917_150 |
0.1723880442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006466_020 |
0.1744315736 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_010931_010 |
0.192867997 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 5 |
Hb_001329_040 |
0.2075479455 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 6 |
Hb_106204_010 |
0.2105531305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_059162_010 |
0.2206015773 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_005218_070 |
0.221165478 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 9 |
Hb_047163_010 |
0.2252231526 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 10 |
Hb_161393_010 |
0.2267202513 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000251_100 |
0.2267662922 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_041769_010 |
0.2290547511 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 13 |
Hb_001173_050 |
0.2291450103 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_002917_020 |
0.2313941677 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_000951_030 |
0.233061575 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000522_040 |
0.2332162259 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 17 |
Hb_002890_400 |
0.2332966023 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 18 |
Hb_183209_010 |
0.2333065981 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002081_040 |
0.2337464491 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 20 |
Hb_004185_040 |
0.2341270827 |
- |
- |
polygalacturonase, putative [Ricinus communis] |