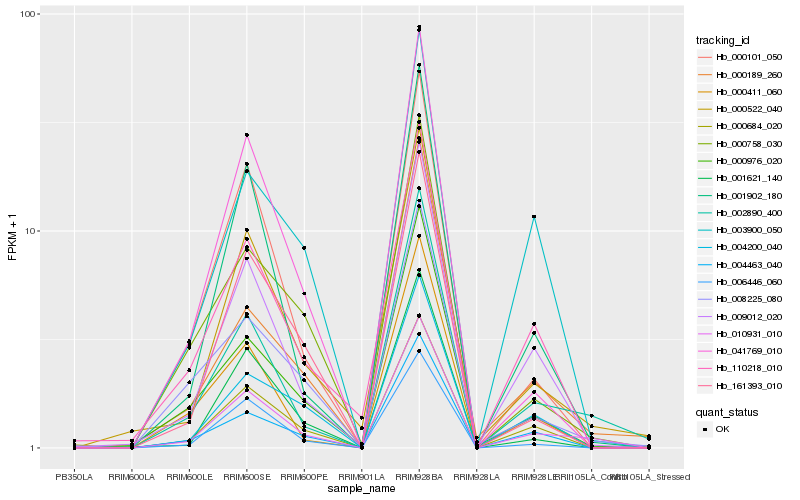
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_010 |
0.0 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 2 |
Hb_002890_400 |
0.0902315284 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 3 |
Hb_009012_020 |
0.1183428109 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 4 |
Hb_000522_040 |
0.1211833777 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 5 |
Hb_004463_040 |
0.1341134496 |
- |
- |
- |
| 6 |
Hb_000684_020 |
0.1348824013 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001902_180 |
0.1352196313 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 8 |
Hb_000976_020 |
0.1401019338 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_006446_060 |
0.1419290474 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 10 |
Hb_008225_080 |
0.1446805367 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 11 |
Hb_000189_260 |
0.1448974909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004200_040 |
0.1449695789 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_000758_030 |
0.1450191581 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000101_050 |
0.1454057575 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_041769_010 |
0.1472895204 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 16 |
Hb_110218_010 |
0.1475358575 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 17 |
Hb_161393_010 |
0.1491556133 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001621_140 |
0.1515220585 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 19 |
Hb_003900_050 |
0.1518062979 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 20 |
Hb_000411_060 |
0.1518703907 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |