| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
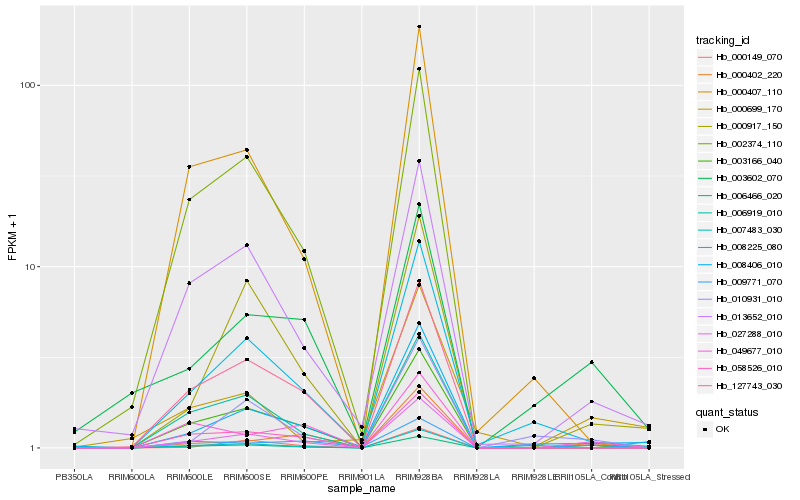
| 1 |
Hb_006466_020 |
0.0 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_000149_070 |
0.1744315736 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009771_070 |
0.2124489194 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 4 |
Hb_013652_010 |
0.2291425593 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_049677_010 |
0.2397589966 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_003602_070 |
0.2453856428 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 7 |
Hb_000402_220 |
0.2502153353 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_007483_030 |
0.2603235368 |
- |
- |
- |
| 9 |
Hb_008406_010 |
0.2621870868 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 10 |
Hb_000699_170 |
0.262382217 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 11 |
Hb_027288_010 |
0.2632307848 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 12 |
Hb_000407_110 |
0.2638609406 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 13 |
Hb_003166_040 |
0.2641778143 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 14 |
Hb_010931_010 |
0.2649909473 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_002374_110 |
0.2668220177 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 16 |
Hb_000917_150 |
0.2671389723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008225_080 |
0.2676285708 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 18 |
Hb_127743_030 |
0.2685318342 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_006919_010 |
0.2686408339 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 20 |
Hb_058526_010 |
0.2698141503 |
- |
- |
- |