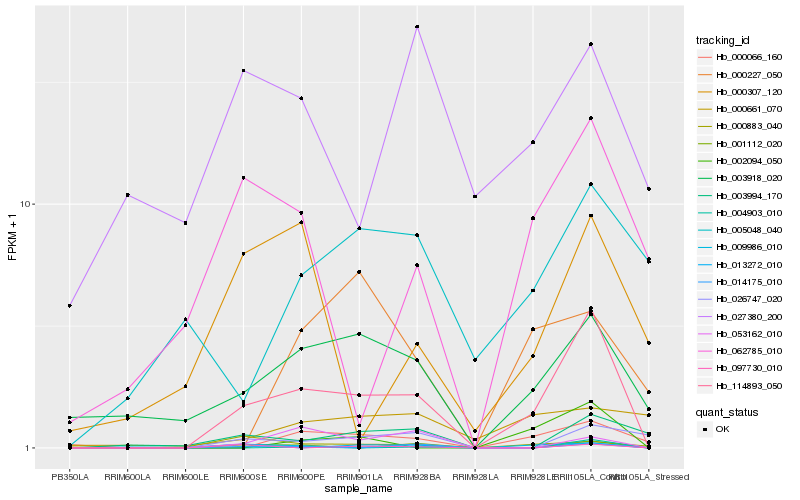
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114893_050 |
0.0 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 2 |
Hb_003918_020 |
0.3261873224 |
- |
- |
- |
| 3 |
Hb_009986_010 |
0.3297621037 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_000066_160 |
0.3416292297 |
- |
- |
- |
| 5 |
Hb_001112_020 |
0.3555248895 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 6 |
Hb_002094_050 |
0.3699944029 |
- |
- |
PREDICTED: organic cation/carnitine transporter 4-like, partial [Eucalyptus grandis] |
| 7 |
Hb_026747_020 |
0.3793044871 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_062785_010 |
0.3797898717 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 9 |
Hb_014175_010 |
0.3824261364 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 10 |
Hb_003994_170 |
0.3842054572 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 11 |
Hb_053162_010 |
0.3909418414 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 12 |
Hb_013272_010 |
0.3957633257 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_000883_040 |
0.3980068314 |
- |
- |
- |
| 14 |
Hb_005048_040 |
0.402781602 |
- |
- |
PREDICTED: endonuclease 1 [Jatropha curcas] |
| 15 |
Hb_097730_010 |
0.4051368365 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 16 |
Hb_000307_120 |
0.4074035341 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 17 |
Hb_000227_050 |
0.4077335258 |
- |
- |
- |
| 18 |
Hb_004903_010 |
0.4146220071 |
- |
- |
- |
| 19 |
Hb_027380_200 |
0.4196380794 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000661_070 |
0.4214340561 |
- |
- |
- |