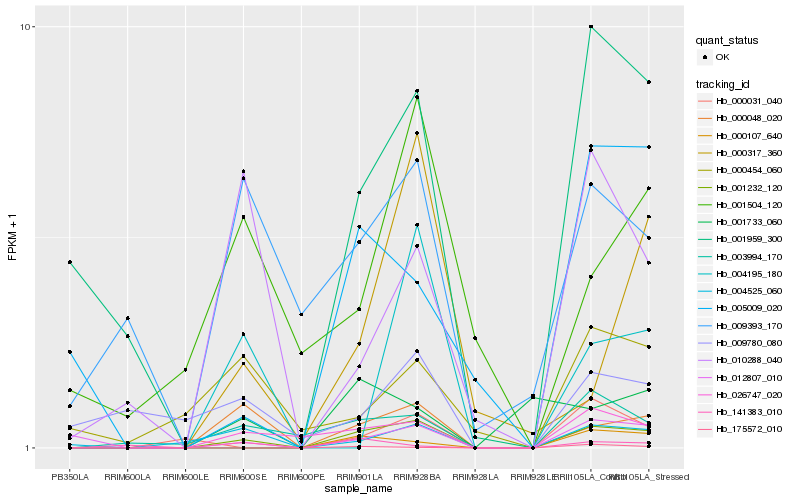
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001232_120 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 2 |
Hb_000048_020 |
0.2276258835 |
- |
- |
- |
| 3 |
Hb_026747_020 |
0.2367264064 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_003994_170 |
0.277527059 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 5 |
Hb_004525_060 |
0.2837923168 |
- |
- |
- |
| 6 |
Hb_012807_010 |
0.2929705153 |
- |
- |
- |
| 7 |
Hb_000107_640 |
0.3093373316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000031_040 |
0.3329884853 |
- |
- |
centromere protein O [Medicago truncatula] |
| 9 |
Hb_001959_300 |
0.3330734543 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 10 |
Hb_009393_170 |
0.3345837706 |
- |
- |
PREDICTED: vestitone reductase-like [Jatropha curcas] |
| 11 |
Hb_005009_020 |
0.3381609474 |
- |
- |
- |
| 12 |
Hb_010288_040 |
0.3395366647 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 13 |
Hb_000317_360 |
0.3435903424 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g08460 [Jatropha curcas] |
| 14 |
Hb_004195_180 |
0.3475542658 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_001504_120 |
0.3564401403 |
- |
- |
- |
| 16 |
Hb_000454_060 |
0.3594081267 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 17 |
Hb_001733_060 |
0.3596684293 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 18 |
Hb_009780_080 |
0.3627227479 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_175572_010 |
0.3656733343 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_141383_010 |
0.3659518272 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |