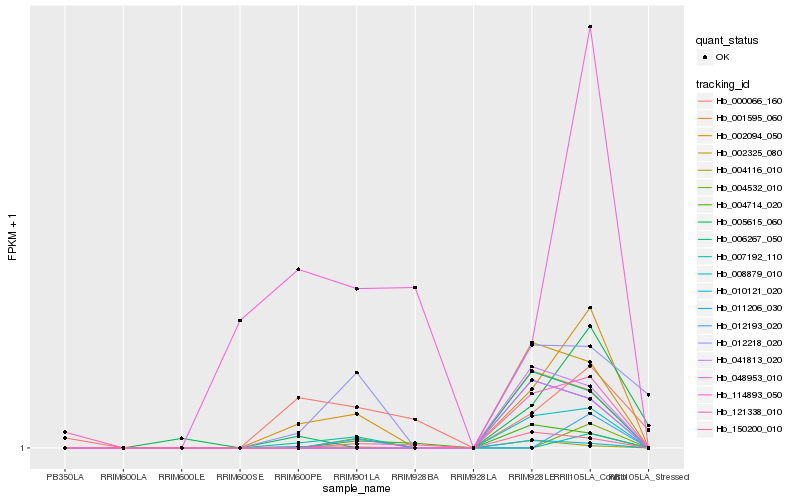
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002094_050 |
0.0 |
- |
- |
PREDICTED: organic cation/carnitine transporter 4-like, partial [Eucalyptus grandis] |
| 2 |
Hb_012218_020 |
0.3574623197 |
- |
- |
hypothetical protein JCGZ_14688 [Jatropha curcas] |
| 3 |
Hb_008879_010 |
0.3607902642 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 4 |
Hb_005615_060 |
0.3638947938 |
- |
- |
PREDICTED: uncharacterized protein LOC105636752 [Jatropha curcas] |
| 5 |
Hb_114893_050 |
0.3699944029 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 6 |
Hb_010121_020 |
0.3788297457 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000066_160 |
0.3918116743 |
- |
- |
- |
| 8 |
Hb_004714_020 |
0.4002610051 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 9 |
Hb_150200_010 |
0.402181779 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 10 |
Hb_004116_010 |
0.405510984 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_002325_080 |
0.4071896709 |
- |
- |
- |
| 12 |
Hb_007192_110 |
0.4092538856 |
- |
- |
- |
| 13 |
Hb_012193_020 |
0.4097265131 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 14 |
Hb_121338_010 |
0.4141888266 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 15 |
Hb_041813_020 |
0.4156572797 |
- |
- |
ATP synthase epsilon subunit [Grubbia rosmarinifolia] |
| 16 |
Hb_001595_060 |
0.4172027144 |
- |
- |
hypothetical protein L484_001429 [Morus notabilis] |
| 17 |
Hb_006267_050 |
0.4174936063 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 18 |
Hb_011206_030 |
0.4205736973 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_048953_010 |
0.4205736973 |
- |
- |
PREDICTED: uncharacterized protein LOC105775428 [Gossypium raimondii] |
| 20 |
Hb_004532_010 |
0.4207570078 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |