| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
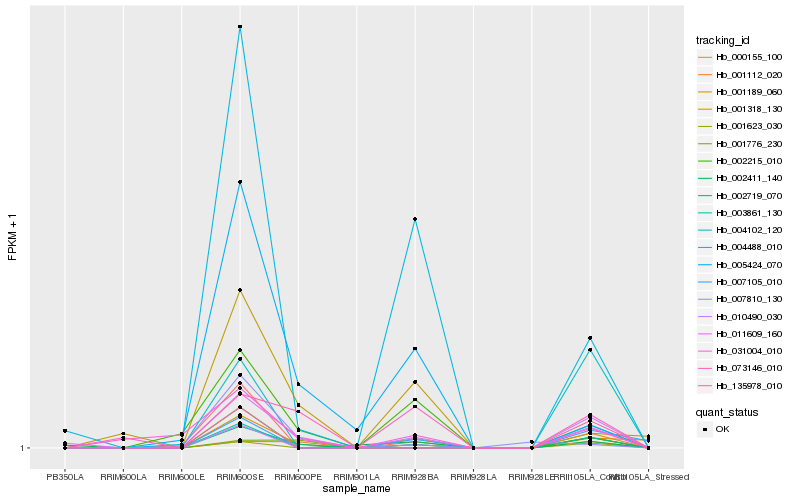
| 1 |
Hb_002719_070 |
0.0 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 2 |
Hb_011609_160 |
0.26337615 |
- |
- |
Lipase, putative [Ricinus communis] |
| 3 |
Hb_001189_060 |
0.2803702396 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 4 |
Hb_010490_030 |
0.3069295186 |
- |
- |
- |
| 5 |
Hb_031004_010 |
0.3102889129 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 6 |
Hb_135978_010 |
0.3114729892 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 7 |
Hb_004488_010 |
0.313083822 |
- |
- |
hypothetical protein CISIN_1g019168mg [Citrus sinensis] |
| 8 |
Hb_073146_010 |
0.3170031409 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 9 |
Hb_001623_030 |
0.3193857181 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_007105_010 |
0.3234981621 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 11 |
Hb_005424_070 |
0.3260149578 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 12 |
Hb_003861_130 |
0.3314838203 |
- |
- |
- |
| 13 |
Hb_000155_100 |
0.345097048 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 14 |
Hb_001318_130 |
0.3469883219 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 15 |
Hb_002215_010 |
0.3495426563 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002411_140 |
0.3496640567 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 17 |
Hb_004102_120 |
0.3500857439 |
- |
- |
- |
| 18 |
Hb_001776_230 |
0.3550301526 |
- |
- |
PREDICTED: uncharacterized protein LOC104884260 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_001112_020 |
0.3561232062 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 20 |
Hb_007810_130 |
0.3598568557 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |