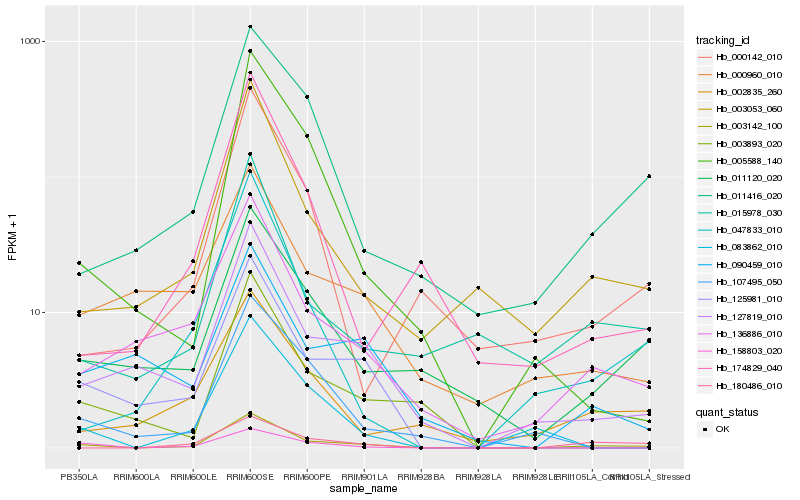
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003142_100 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Eucalyptus grandis] |
| 2 |
Hb_127819_010 |
0.1991101797 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_003053_060 |
0.2026977627 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 4 |
Hb_136886_010 |
0.2080479431 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_011120_020 |
0.2115876433 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_011416_020 |
0.2195403006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_015978_030 |
0.2256228922 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 8 |
Hb_125981_010 |
0.2262143009 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 9 |
Hb_180486_010 |
0.2324617676 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_005588_140 |
0.2346773614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002835_260 |
0.2353952645 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 12 |
Hb_047833_010 |
0.2369207364 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 13 |
Hb_090459_010 |
0.2397084149 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 14 |
Hb_000142_010 |
0.2397740639 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 15 |
Hb_083862_010 |
0.2411509292 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 16 |
Hb_003893_020 |
0.2431268958 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_174829_040 |
0.2436226988 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 18 |
Hb_158803_020 |
0.2440150461 |
- |
- |
hypothetical protein JCGZ_12890 [Jatropha curcas] |
| 19 |
Hb_107495_050 |
0.2459329222 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_000960_010 |
0.2460234721 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |