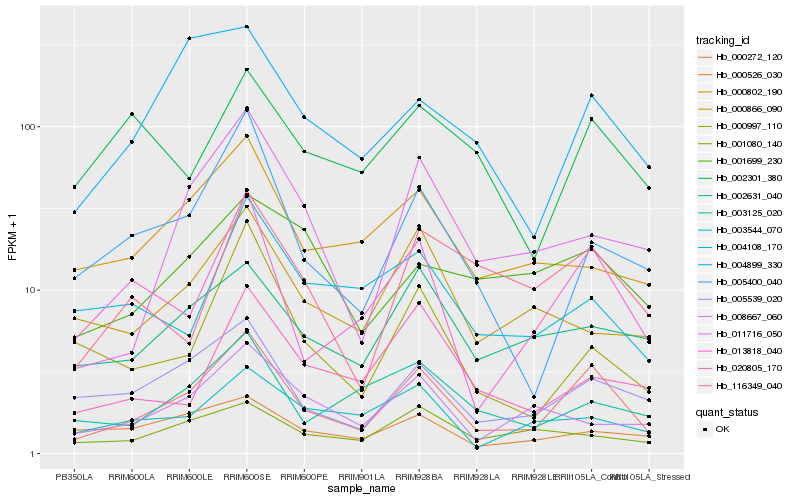
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000272_120 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 2 |
Hb_013818_040 |
0.1738433644 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 3 |
Hb_005539_020 |
0.1827719038 |
- |
- |
- |
| 4 |
Hb_005400_040 |
0.2026522209 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 5 |
Hb_002631_040 |
0.2052525363 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 6 |
Hb_011716_050 |
0.2084260707 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 7 |
Hb_004899_330 |
0.2107249931 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 8 |
Hb_003125_020 |
0.2147168093 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_000802_190 |
0.2174585603 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 10 |
Hb_116349_040 |
0.2175818906 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 11 |
Hb_000997_110 |
0.2181533219 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 12 |
Hb_003544_070 |
0.2190576665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001080_140 |
0.2210717715 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_020805_170 |
0.2223499054 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 15 |
Hb_008667_060 |
0.2252378 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 16 |
Hb_000866_090 |
0.2279806242 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_001699_230 |
0.2289924285 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_004108_170 |
0.2293189509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002301_380 |
0.2321720796 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_000526_030 |
0.232486065 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |