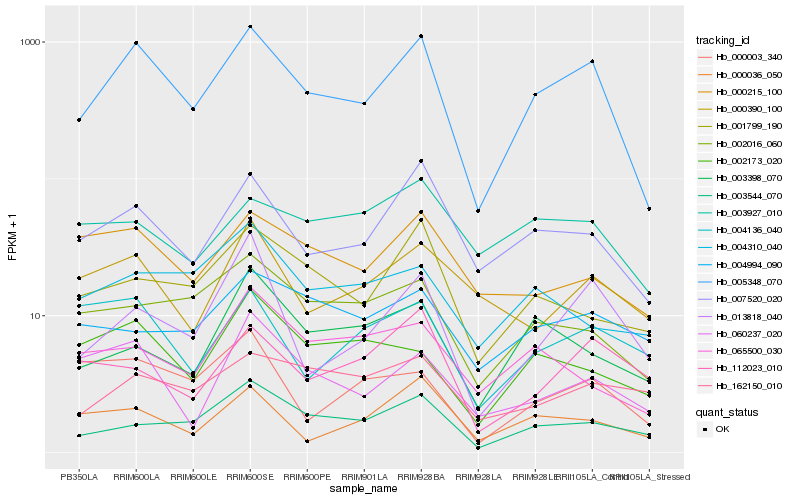
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005348_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007520_020 |
0.150127242 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |
| 3 |
Hb_003544_070 |
0.1805871263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002016_060 |
0.1853032984 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 5 |
Hb_001799_190 |
0.1916066723 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 6 |
Hb_000036_050 |
0.1935825087 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 7 |
Hb_000215_100 |
0.1949729695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_013818_040 |
0.1953675271 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 9 |
Hb_060237_020 |
0.2001755186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000390_100 |
0.2043687221 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 11 |
Hb_002173_020 |
0.205781402 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 12 |
Hb_003927_010 |
0.2061755128 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_004136_040 |
0.2064399616 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 14 |
Hb_162150_010 |
0.2064988911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000003_340 |
0.2074930957 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_003398_070 |
0.2083844023 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 17 |
Hb_004310_040 |
0.2112056522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_112023_010 |
0.2117674582 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 19 |
Hb_004994_090 |
0.2128896375 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 20 |
Hb_065500_030 |
0.214116827 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |