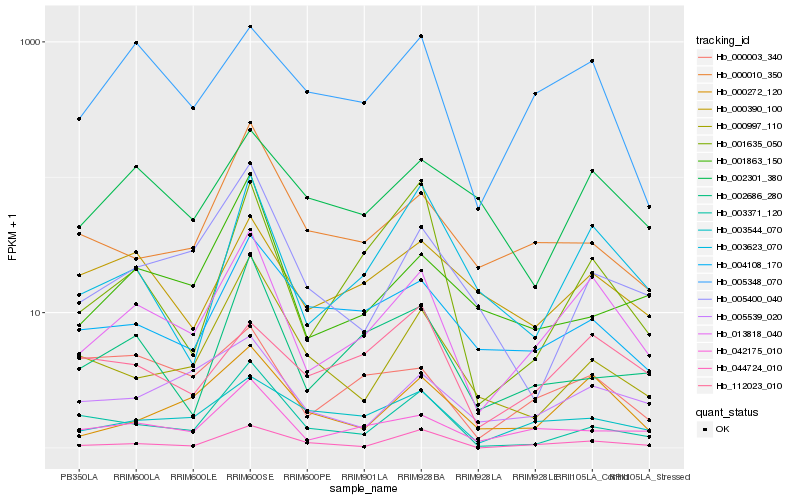
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013818_040 |
0.0 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 2 |
Hb_042175_010 |
0.1737889744 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 3 |
Hb_000272_120 |
0.1738433644 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003623_070 |
0.1900561806 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_005348_070 |
0.1953675271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_044724_010 |
0.1979410621 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 7 |
Hb_003544_070 |
0.2014361277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005539_020 |
0.2056176153 |
- |
- |
- |
| 9 |
Hb_004108_170 |
0.2096173812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001635_050 |
0.2176024583 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Nicotiana sylvestris] |
| 11 |
Hb_003371_120 |
0.217830085 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 12 |
Hb_002686_280 |
0.2180562333 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_112023_010 |
0.2206040776 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 14 |
Hb_000390_100 |
0.2207610049 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 15 |
Hb_000010_350 |
0.2231484399 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 16 |
Hb_000003_340 |
0.224422251 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_005400_040 |
0.2245598303 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 18 |
Hb_002301_380 |
0.2247245751 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_001863_150 |
0.2265899983 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 20 |
Hb_000997_110 |
0.2296282673 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |