| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
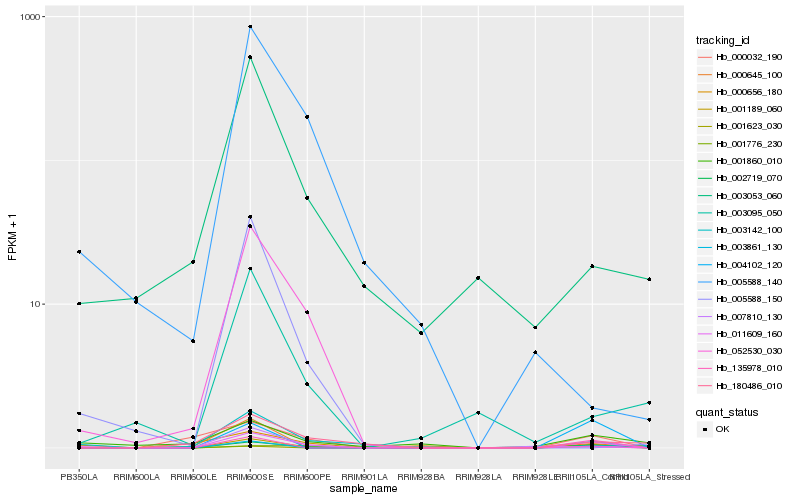
Hb_011609_160 |
0.0 |
- |
- |
Lipase, putative [Ricinus communis] |
| 2 |
Hb_002719_070 |
0.26337615 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 3 |
Hb_135978_010 |
0.3134983372 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 4 |
Hb_001623_030 |
0.3269875729 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_003142_100 |
0.3301349854 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Eucalyptus grandis] |
| 6 |
Hb_000656_180 |
0.3446153468 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 7 |
Hb_003861_130 |
0.3446345137 |
- |
- |
- |
| 8 |
Hb_000032_190 |
0.3449738652 |
- |
- |
- |
| 9 |
Hb_001776_230 |
0.3480458168 |
- |
- |
PREDICTED: uncharacterized protein LOC104884260 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_001189_060 |
0.3521593044 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 11 |
Hb_000645_100 |
0.3544302651 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 12 |
Hb_007810_130 |
0.3650884011 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 13 |
Hb_001860_010 |
0.367819655 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_003053_060 |
0.3684167243 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 15 |
Hb_004102_120 |
0.3689922402 |
- |
- |
- |
| 16 |
Hb_180486_010 |
0.3697465245 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_052530_030 |
0.3699501534 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 18 |
Hb_005588_140 |
0.3727743179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003095_050 |
0.3758133451 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 20 |
Hb_005588_150 |
0.376769232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |