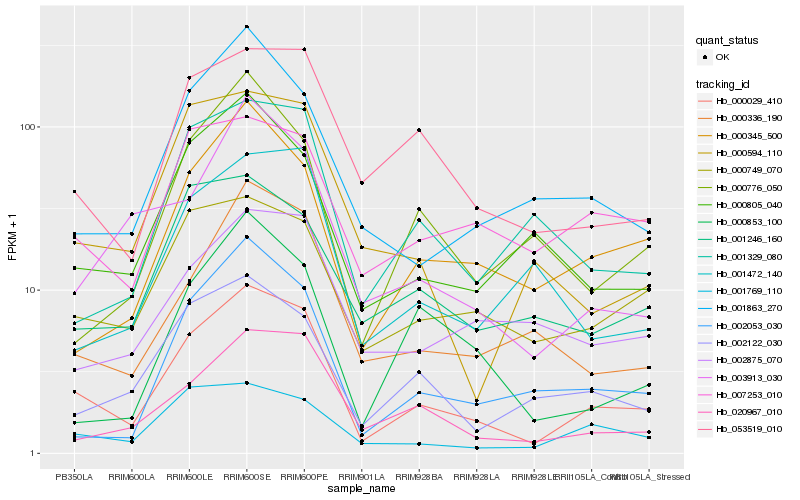
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_410 |
0.0 |
- |
- |
AT14A, putative [Ricinus communis] |
| 2 |
Hb_020967_010 |
0.1611934824 |
- |
- |
- |
| 3 |
Hb_053519_010 |
0.1701943604 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 4 |
Hb_000336_190 |
0.172134418 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 5 |
Hb_002053_030 |
0.1744135421 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000345_500 |
0.1747398321 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 7 |
Hb_002122_030 |
0.1754581483 |
- |
- |
- |
| 8 |
Hb_001769_110 |
0.1757203368 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 9 |
Hb_000805_040 |
0.1772384013 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 10 |
Hb_001863_270 |
0.1773957559 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 11 |
Hb_000749_070 |
0.1783354973 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 12 |
Hb_003913_030 |
0.190887152 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 13 |
Hb_001329_080 |
0.1910319162 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 14 |
Hb_007253_010 |
0.1912551397 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_000594_110 |
0.1912607951 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 16 |
Hb_001246_160 |
0.1937935728 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 17 |
Hb_002875_070 |
0.1962852836 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 18 |
Hb_001472_140 |
0.1966284257 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 19 |
Hb_000776_050 |
0.197253028 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_000853_100 |
0.1977317507 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |