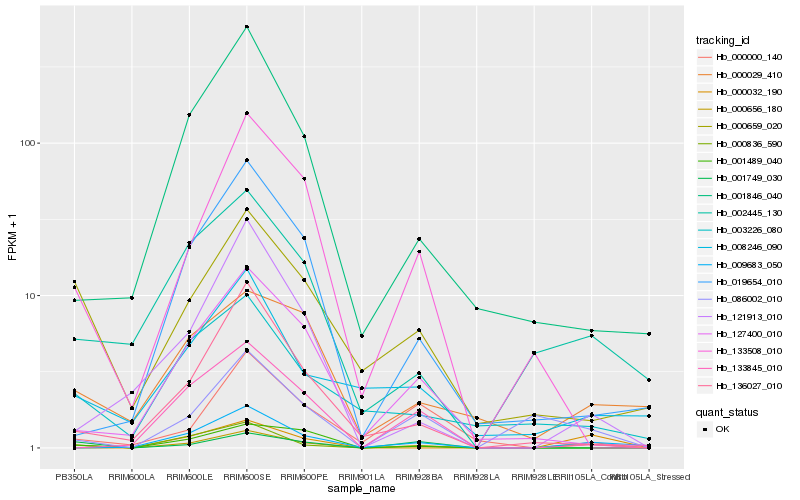
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 2 |
Hb_009683_050 |
0.248110017 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 3 |
Hb_002445_130 |
0.2666058612 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 4 |
Hb_086002_010 |
0.2703894737 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_000659_020 |
0.2726445187 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 6 |
Hb_133508_010 |
0.2740716862 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 7 |
Hb_000029_410 |
0.2808052067 |
- |
- |
AT14A, putative [Ricinus communis] |
| 8 |
Hb_127400_010 |
0.281438163 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000836_590 |
0.2829562669 |
- |
- |
hypothetical protein JCGZ_14318 [Jatropha curcas] |
| 10 |
Hb_136027_010 |
0.2833883727 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_003226_080 |
0.2836734487 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 12 |
Hb_008246_090 |
0.2864358658 |
- |
- |
- |
| 13 |
Hb_001749_030 |
0.2884423193 |
- |
- |
- |
| 14 |
Hb_001489_040 |
0.2892864722 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 15 |
Hb_000000_140 |
0.2903842887 |
- |
- |
- |
| 16 |
Hb_000032_190 |
0.2921282621 |
- |
- |
- |
| 17 |
Hb_001846_040 |
0.2942579822 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_121913_010 |
0.2951488997 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_133845_010 |
0.2961574426 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_019654_010 |
0.2961913811 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |